
| HUGE |
Gene/Protein Characteristic Table for KIAA1496 |
|
Link to :
GTOP | SWISS-PROT/TrEMBL | GeneCards| RefDIC | |
| Features : DNA sequence | Protein sequence | Expression | Mapping | |
| Product ID : | ORK04595 |
|---|---|
| Accession No. : | AB040929 |
| Description : | Contactin-3 precursor. |
| HUGO Gene Name : | |
| Clone Name : | fj09513 [Vector Info] |
| Flexi ORF Clone : | pF1KA1496
 |
| Source : | Human fetal brain |
Features of the cloned DNA sequence |
Description | |
|---|---|---|
Length: 4594 bp
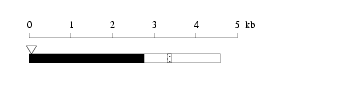
|
| cloned DNA seq. | |
Warning for N-terminal truncation: | YES |
Warning for coding interruption: | NO |
Length of 3'UTR 1830 bp Genome contig ID gi89161205r_74294412 PolyA signal sequence
(AATAAA,-19)
TATTATTTTCTATTCTAATAAAATTTATAACAAGCFlanking genome sequence
(100000 - 99951)
AAATGAGTGTGCAGTATTAATTGCGGTATGGTCAAAGAAGAATGTCTACA
Chr f/r start end exon identity class ContigView(URL based/DAS) 3 r 74394412 74556781 19 99.9 Terminal No-hit
Features of the protein sequence |
Description | |
|---|---|---|
Length: 920 aa
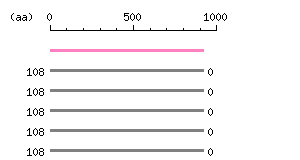 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
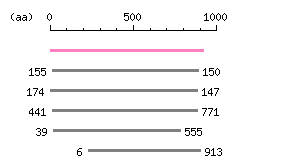 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.

Expression profile |
Description | |
|---|---|---|
| RT-PCR-ELISA | Description |
|---|

Experimental conditions
| : GGAATGCCACAGACACTAAAG | |
| : GTCCTCTTTAATGGGCAGCAC | |
| : 95 °C |
RH mapping information |
Description | |
|---|---|---|
| : 3 |
| : unigene | |
| : - | |
| : - | |
| : - | |
| : - |
|
How to obtain KIAA clone(s) Back to the HUGE Protein Database homepage 
| |