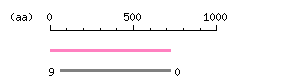
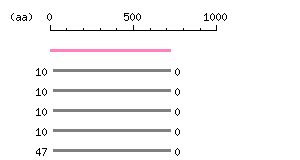Length: 6247 bp
|
| cloned DNA seq. | |
Warning for N-terminal truncation: | NO |
Warning for coding interruption: | NO |
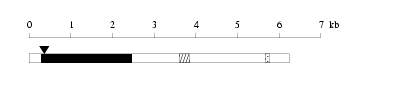
Length of 3'UTR 3775 bp Genome contig ID gi65511124r_78000196 PolyA signal sequence
(AATAAA,-22)
AAATTTTCATTACAATAAAAAATGACTTTGCTCTGFlanking genome sequence
(100000 - 99951)
AAAGTGAGTGCCTTGGACTGGCTTGGTAATTGATCAACTTGAGGTATTTG
KIAA Alignment based on: KIAA0307 DNA sequence, AA sequence, Physical map