|
Order Kazusa clone(s) from : |
|
Order Kazusa clone(s) from : |
| Product ID | ORK00598 |
|---|---|
| Accession No | AB014565 |
| Description | RAB11 family interacting protein 3 (class II), transcript variant 1 |
| Clone name | hk02064 |
| Vector information | |
| cDNA sequence | DNA sequence (4080 bp) Predicted protein sequence (760 aa) |
|
HaloTag ORF Clone |
FHC00598

|
| Flexi ORF Clone | FXC00598 |
| Source | Human adult brain |
| Rouge ID |
mKIAA0665
by Kazusa Mouse cDNA Project
|
 Length: 4080 bp
Length: 4080 bp Physical map
Physical map
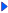 Restriction map
Restriction map Prediction of protein coding region (GeneMark analysis).
Prediction of protein coding region (GeneMark analysis).
| N-terminal truncation | Coding interruption | |
|---|---|---|
| cloned DNA seq | No warning | No warning |
 Length: 760 aa
Length: 760 aa Result of homology search against nr database
(FASTA output,
Multiple alignment)
Result of homology search against nr database
(FASTA output,
Multiple alignment)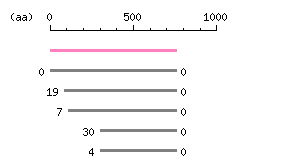 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
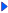 Result of homology search against HUGE database
(FASTA output,
Multiple alignment)
Result of homology search against HUGE database
(FASTA output,
Multiple alignment)The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
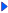 Result of motif / domain search (InterProScan and SOSUI)
Result of motif / domain search (InterProScan and SOSUI)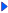 Result of InterProScan
Result of InterProScan
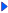 RT-PCR
RT-PCR
|
|---|
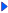 RT-PCR-ELISA
RT-PCR-ELISA
|
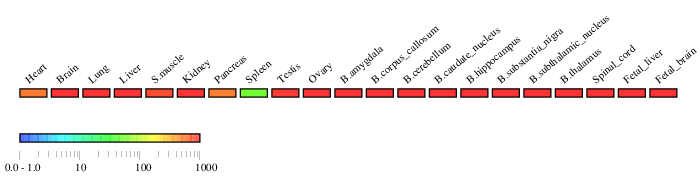
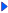 Experimental conditions
Experimental conditions| Primer_f | ACCTGAGATGACCGCTGTGTG |
|---|---|
| Primer_r | CGATGTTTGGATTCTGGCCTG |
| PCR conditions | 95 °C 30 sec 30 sec 55 °C 55 °C 30 sec 30 sec 72 °C 72 °C 60 sec 60 sec 30 cycles 30 cycles |
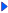 Chromosome No. 16
Chromosome No. 16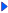 Experimental conditions
Experimental conditions| Panel name | GeneBridge 4 |
|---|---|
| Primer_f | ACCTGAGATGACCGCTGTGTG |
| Primer_r | CGATGTTTGGATTCTGGCCTG |
| PCR product length | 159 bp |
| PCR conditions | 95 °C 15 sec 15 sec 64 °C 64 °C 60 sec 60 sec 30 cycles 30 cycles |