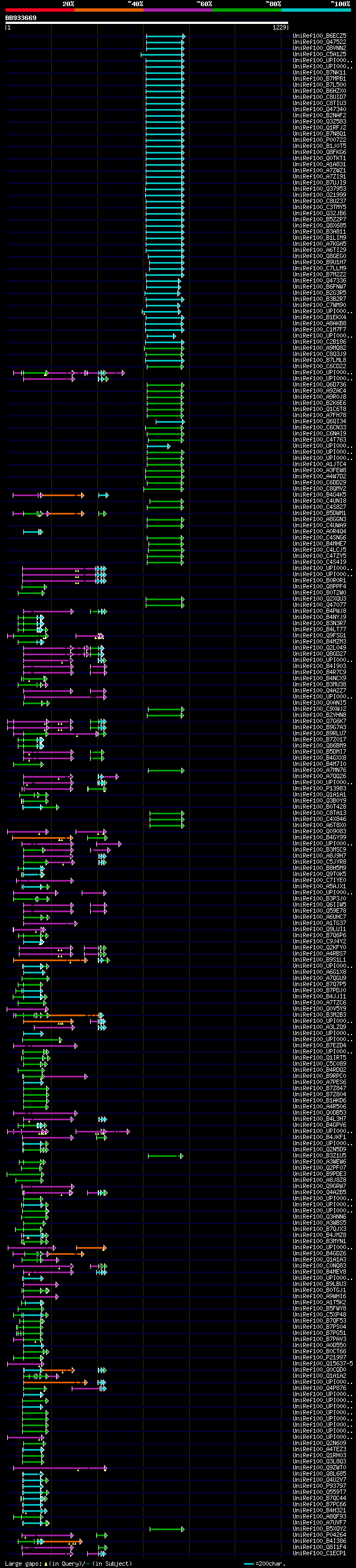
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB933669 RCC06178
(1229 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B6ECZ5 LacZ alpha n=1 Tax=Cloning vector pMAK28 RepID=... 117 3e-24
UniRef100_Q47522 LacZ protein (Fragment) n=1 Tax=Escherichia col... 114 2e-23
UniRef100_Q8VNN2 Beta-galactosidase n=1 Tax=Escherichia coli Rep... 114 2e-23
UniRef100_C5A125 Tryptophan synthase alpha chain n=1 Tax=Escheri... 114 2e-23
UniRef100_UPI0001B5303F beta-D-galactosidase n=1 Tax=Shigella sp... 113 5e-23
UniRef100_UPI0001B525AD beta-D-galactosidase n=1 Tax=Escherichia... 113 5e-23
UniRef100_B7NK11 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 113 5e-23
UniRef100_B7MPB1 Beta-D-galactosidase n=1 Tax=Escherichia coli E... 113 5e-23
UniRef100_B7L500 Beta-D-galactosidase n=1 Tax=Escherichia coli 5... 113 5e-23
UniRef100_B6HZX0 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 113 5e-23
UniRef100_C8UID7 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 113 5e-23
UniRef100_C8TIU3 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 113 5e-23
UniRef100_Q47340 LacZ 5'-region (Fragment) n=2 Tax=Escherichia c... 113 5e-23
UniRef100_B2NAF2 Beta-galactosidase n=1 Tax=Escherichia coli 536... 113 5e-23
UniRef100_Q3Z583 Beta-galactosidase n=1 Tax=Shigella sonnei Ss04... 113 5e-23
UniRef100_Q1RFJ2 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 113 5e-23
UniRef100_B7N8Q1 Beta-galactosidase n=1 Tax=Escherichia coli UMN... 113 5e-23
UniRef100_P00722 Beta-galactosidase n=6 Tax=Escherichia coli Rep... 113 5e-23
UniRef100_B1J0T5 Beta-galactosidase n=1 Tax=Escherichia coli ATC... 113 5e-23
UniRef100_Q8FKG6 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 113 5e-23
UniRef100_Q0TKT1 Beta-galactosidase n=1 Tax=Escherichia coli 536... 113 5e-23
UniRef100_A1A831 Beta-galactosidase n=1 Tax=Escherichia coli APE... 113 5e-23
UniRef100_A7ZWZ1 Beta-galactosidase n=1 Tax=Escherichia coli HS ... 113 5e-23
UniRef100_A7ZI91 Beta-galactosidase n=1 Tax=Escherichia coli E24... 113 5e-23
UniRef100_B7UJI9 Beta-galactosidase n=1 Tax=Escherichia coli O12... 112 6e-23
UniRef100_Q37953 LacZ protein (Fragment) n=1 Tax=Phage M13mp18 R... 112 1e-22
UniRef100_O21999 LacZ-alpha n=1 Tax=Cloning vector pAL-Z RepID=O... 112 1e-22
UniRef100_C8U237 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 111 1e-22
UniRef100_C3TMY5 Beta-D-galactosidase n=1 Tax=Escherichia coli R... 110 2e-22
UniRef100_Q32JB6 Beta-galactosidase n=1 Tax=Shigella dysenteriae... 110 2e-22
UniRef100_B5Z2P7 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 110 2e-22
UniRef100_Q8X685 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 110 2e-22
UniRef100_B3A811 Beta-galactosidase n=1 Tax=Escherichia coli O15... 109 5e-22
UniRef100_B1LIM9 Beta-galactosidase n=1 Tax=Escherichia coli SMS... 109 5e-22
UniRef100_A7KGA5 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 109 5e-22
UniRef100_A6TI29 Beta-galactosidase 2 n=1 Tax=Klebsiella pneumon... 109 5e-22
UniRef100_Q8GEG0 Putative uncharacterized protein n=1 Tax=Erwini... 108 9e-22
UniRef100_B9U1H7 Beta-D-galactosidase n=1 Tax=Vaccinia virus GLV... 107 2e-21
UniRef100_C7LLM9 Beta-galactosidase, chain D n=1 Tax=Mycoplasma ... 107 2e-21
UniRef100_B7M2Z2 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 107 4e-21
UniRef100_Q47336 LacZ-alpha peptide n=1 Tax=Escherichia coli Rep... 103 5e-20
UniRef100_B6FNW7 Putative uncharacterized protein n=1 Tax=Clostr... 103 5e-20
UniRef100_B2G3R5 Putative puroindoline b protein n=1 Tax=Triticu... 102 7e-20
UniRef100_B3B2R7 Beta-galactosidase n=1 Tax=Escherichia coli O15... 102 1e-19
UniRef100_C7WM90 LacZ alpha protein (Fragment) n=1 Tax=Enterococ... 101 1e-19
UniRef100_UPI0000ECC20B UPI0000ECC20B related cluster n=1 Tax=Ga... 100 3e-19
UniRef100_B1EKX4 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 93 5e-17
UniRef100_A8AKB8 Beta-galactosidase n=1 Tax=Citrobacter koseri A... 90 6e-16
UniRef100_C1M7F7 Beta-D-galactosidase n=1 Tax=Citrobacter sp. 30... 89 8e-16
UniRef100_UPI0000ECBAF8 UPI0000ECBAF8 related cluster n=1 Tax=Ga... 87 4e-15
UniRef100_C2B186 Putative uncharacterized protein n=1 Tax=Citrob... 87 4e-15
UniRef100_A9MQ82 Beta-galactosidase n=1 Tax=Salmonella enterica ... 82 2e-13
UniRef100_C8Q3J9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 81 3e-13
UniRef100_B7LML8 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 78 2e-12
UniRef100_C6CD22 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 77 5e-12
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 64 7e-12
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 55 2e-11
UniRef100_Q6D736 Beta-galactosidase n=1 Tax=Pectobacterium atros... 74 3e-11
UniRef100_A9ZAC4 Beta-galactosidase (Lactase) n=2 Tax=Yersinia p... 73 6e-11
UniRef100_A9R0J8 Beta-galactosidase n=6 Tax=Yersinia pestis RepI... 73 6e-11
UniRef100_B2K6E6 Beta-galactosidase n=3 Tax=Yersinia pseudotuber... 73 6e-11
UniRef100_Q1C6T8 Beta-galactosidase n=8 Tax=Yersinia pestis RepI... 73 6e-11
UniRef100_A7FH78 Beta-galactosidase n=1 Tax=Yersinia pseudotuber... 73 6e-11
UniRef100_Q6QI34 LRRGT00174 n=1 Tax=Rattus norvegicus RepID=Q6QI... 72 1e-10
UniRef100_C6CN33 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 1e-10
UniRef100_C6NAI9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 1e-10
UniRef100_C4T763 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 1e-10
UniRef100_UPI0000ECAFAB UPI0000ECAFAB related cluster n=1 Tax=Ga... 72 2e-10
UniRef100_UPI0001826C8D hypothetical protein ENTCAN_01162 n=1 Ta... 71 2e-10
UniRef100_UPI0001A43550 beta-D-galactosidase n=1 Tax=Pectobacter... 71 3e-10
UniRef100_A1JTC4 Beta-galactosidase n=1 Tax=Yersinia enterocolit... 71 3e-10
UniRef100_A3FEW8 Beta-galactosidase n=1 Tax=Pantoea agglomerans ... 71 3e-10
UniRef100_A4W7D2 Beta-galactosidase n=1 Tax=Enterobacter sp. 638... 71 3e-10
UniRef100_C6DD29 Beta-galactosidase n=1 Tax=Pectobacterium carot... 70 4e-10
UniRef100_C8QMV2 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 4e-10
UniRef100_B4G4K5 GL24546 n=1 Tax=Drosophila persimilis RepID=B4G... 40 4e-10
UniRef100_C4UNI8 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 6e-10
UniRef100_C4S827 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 6e-10
UniRef100_B5DWM1 GA26446 n=1 Tax=Drosophila pseudoobscura pseudo... 52 8e-10
UniRef100_A8GGN3 Beta-galactosidase n=1 Tax=Serratia proteamacul... 69 1e-09
UniRef100_C4UWA9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 68 2e-09
UniRef100_A0R4Q4 Putative uncharacterized protein n=1 Tax=Mycoba... 67 3e-09
UniRef100_C4SNG6 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 3e-09
UniRef100_B4MHE7 GJ14580 n=1 Tax=Drosophila virilis RepID=B4MHE7... 67 4e-09
UniRef100_C4LCJ5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 5e-09
UniRef100_C4TZY5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 5e-09
UniRef100_C4S4I9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 5e-09
UniRef100_UPI000175F21B PREDICTED: similar to Ras-associated and... 52 6e-09
UniRef100_UPI00015A6C38 UPI00015A6C38 related cluster n=1 Tax=Da... 52 6e-09
UniRef100_B0R0R1 Novel protein similar to human Ras association ... 52 6e-09
UniRef100_Q8PPF4 Putative uncharacterized protein n=1 Tax=Xantho... 66 7e-09
UniRef100_B0T2W0 OmpA/MotB domain protein n=1 Tax=Caulobacter sp... 66 7e-09
UniRef100_Q2XQU3 Beta-galactosidase 2 n=1 Tax=Enterobacter cloac... 66 7e-09
UniRef100_Q47077 Beta-galactosidase n=1 Tax=Enterobacter cloacae... 66 7e-09
UniRef100_B4PWJ8 GE16594 n=1 Tax=Drosophila yakuba RepID=B4PWJ8_... 58 9e-09
UniRef100_B4NYJ9 GE18286 n=1 Tax=Drosophila yakuba RepID=B4NYJ9_... 58 1e-08
UniRef100_B3N3R7 GG25000 n=1 Tax=Drosophila erecta RepID=B3N3R7_... 58 1e-08
UniRef100_B4LT77 GJ19953 n=1 Tax=Drosophila virilis RepID=B4LT77... 56 1e-08
UniRef100_Q9FSG1 Putative extensin n=1 Tax=Nicotiana sylvestris ... 56 1e-08
UniRef100_B4MZM3 GK24347 n=1 Tax=Drosophila willistoni RepID=B4M... 54 2e-08
UniRef100_Q2L049 Filamentous hemagglutinin/adhesin n=1 Tax=Borde... 52 2e-08
UniRef100_Q8GD27 Adhesin FhaB n=1 Tax=Bordetella avium RepID=Q8G... 52 2e-08
UniRef100_UPI0000EBDEB7 PREDICTED: similar to formin-like 1 n=1 ... 48 2e-08
UniRef100_B4I903 GM19047 n=1 Tax=Drosophila sechellia RepID=B4I9... 55 2e-08
UniRef100_B4R7C9 GD16486 n=1 Tax=Drosophila simulans RepID=B4R7C... 55 2e-08
UniRef100_B4NCX9 GK10119 n=1 Tax=Drosophila willistoni RepID=B4N... 64 3e-08
UniRef100_B3MU38 GF21178 n=1 Tax=Drosophila ananassae RepID=B3MU... 56 3e-08
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 57 3e-08
UniRef100_UPI0000D99778 PREDICTED: hypothetical protein n=1 Tax=... 64 3e-08
UniRef100_Q0ANI5 OmpA/MotB domain protein n=1 Tax=Maricaulis mar... 64 3e-08
UniRef100_C9XWJ2 Beta-galactosidase n=1 Tax=Cronobacter turicens... 64 3e-08
UniRef100_B2VHN8 Beta-galactosidase n=1 Tax=Erwinia tasmaniensis... 64 3e-08
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 58 4e-08
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 58 4e-08
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 56 4e-08
UniRef100_B7Z017 CG33003, isoform B n=1 Tax=Drosophila melanogas... 56 4e-08
UniRef100_Q86BM9 CG33003, isoform A n=1 Tax=Drosophila melanogas... 56 4e-08
UniRef100_B5DMI7 GA26754 n=1 Tax=Drosophila pseudoobscura pseudo... 56 4e-08
UniRef100_B4GXX8 GL20180 n=1 Tax=Drosophila persimilis RepID=B4G... 56 4e-08
UniRef100_B4M7I0 GJ16994 n=1 Tax=Drosophila virilis RepID=B4M7I0... 64 4e-08
UniRef100_A7MN76 Beta-galactosidase n=1 Tax=Cronobacter sakazaki... 63 6e-08
UniRef100_A7QQ26 Chromosome chr2 scaffold_140, whole genome shot... 44 6e-08
UniRef100_UPI0000E4981C PREDICTED: hypothetical protein n=1 Tax=... 51 6e-08
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 56 7e-08
UniRef100_Q1A1A1 Forkhead box protein G1 n=1 Tax=Ceratotherium s... 44 7e-08
UniRef100_Q3B0Y9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 63 8e-08
UniRef100_B0T428 Glycoside hydrolase family 16 n=1 Tax=Caulobact... 63 8e-08
UniRef100_C8TA13 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 63 8e-08
UniRef100_C4X846 Beta-D-galactosidase n=1 Tax=Klebsiella pneumon... 63 8e-08
UniRef100_A6T8X0 Beta-galactosidase 1 n=1 Tax=Klebsiella pneumon... 63 8e-08
UniRef100_Q09083 Hydroxyproline-rich glycoprotein n=1 Tax=Phaseo... 55 9e-08
UniRef100_B4GY99 GL19880 n=1 Tax=Drosophila persimilis RepID=B4G... 46 9e-08
UniRef100_UPI0001982DB4 PREDICTED: hypothetical protein n=1 Tax=... 55 9e-08
UniRef100_B3MSC9 GF20819 n=1 Tax=Drosophila ananassae RepID=B3MS... 56 9e-08
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 53 1e-07
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 57 1e-07
UniRef100_B8H5M9 Cell surface antigen Sca2 n=2 Tax=Caulobacter v... 62 1e-07
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 62 1e-07
UniRef100_C7IYE0 Os02g0131200 protein (Fragment) n=1 Tax=Oryza s... 62 1e-07
UniRef100_A5AJX1 Putative uncharacterized protein n=1 Tax=Vitis ... 62 1e-07
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 52 1e-07
UniRef100_B3P3J0 GG21917 n=1 Tax=Drosophila erecta RepID=B3P3J0_... 42 1e-07
UniRef100_Q6IIW5 HDC16773 n=1 Tax=Drosophila melanogaster RepID=... 53 1e-07
UniRef100_Q59E78 CG13376 n=1 Tax=Drosophila melanogaster RepID=Q... 53 1e-07
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 62 1e-07
UniRef100_A1TG37 Putative uncharacterized protein n=1 Tax=Mycoba... 62 1e-07
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 62 1e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 62 1e-07
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 62 1e-07
UniRef100_Q2KFY0 Putative uncharacterized protein n=1 Tax=Magnap... 51 1e-07
UniRef100_A4RBS7 Putative uncharacterized protein n=1 Tax=Magnap... 51 1e-07
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 46 2e-07
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 62 2e-07
UniRef100_A6G1X8 Single-stranded DNA-binding protein n=1 Tax=Ple... 62 2e-07
UniRef100_A7QGU9 Chromosome chr16 scaffold_94, whole genome shot... 62 2e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 62 2e-07
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 62 2e-07
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 62 2e-07
UniRef100_A7TZC6 Putative uncharacterized protein n=1 Tax=Lepeop... 62 2e-07
UniRef100_Q0V5Y9 Putative uncharacterized protein n=1 Tax=Phaeos... 62 2e-07
UniRef100_B3M2B3 GF17078 n=1 Tax=Drosophila ananassae RepID=B3M2... 57 2e-07
UniRef100_UPI000186EF1C formin 1,2/cappuccino, putative n=1 Tax=... 41 2e-07
UniRef100_A3LZQ9 Predicted protein n=1 Tax=Pichia stipitis RepID... 45 2e-07
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 61 2e-07
UniRef100_UPI0000E4896A PREDICTED: similar to CG33556-PA n=1 Tax... 61 2e-07
UniRef100_B7EZD4 cDNA clone:002-104-C03, full insert sequence n=... 61 2e-07
UniRef100_UPI0000D9F56F PREDICTED: similar to Protein CXorf45 n=... 61 2e-07
UniRef100_Q1IRT5 Putative uncharacterized protein n=1 Tax=Candid... 61 2e-07
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 61 2e-07
UniRef100_B4RDQ2 OmpA family protein n=1 Tax=Phenylobacterium zu... 61 2e-07
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 61 2e-07
UniRef100_A7PES6 Chromosome chr11 scaffold_13, whole genome shot... 61 2e-07
UniRef100_B7Z847 cDNA FLJ52583 n=1 Tax=Homo sapiens RepID=B7Z847... 61 2e-07
UniRef100_B7Z804 cDNA FLJ61626 n=1 Tax=Homo sapiens RepID=B7Z804... 61 2e-07
UniRef100_B1AKD6 Asparagine-linked glycosylation 13 homolog (S. ... 61 2e-07
UniRef100_A4R506 Putative uncharacterized protein n=1 Tax=Magnap... 61 2e-07
UniRef100_Q0DB53 DEAD-box ATP-dependent RNA helicase 52A n=2 Tax... 61 2e-07
UniRef100_B4L3H7 GI15551 n=1 Tax=Drosophila mojavensis RepID=B4L... 53 2e-07
UniRef100_B4GPV6 GL15122 n=1 Tax=Drosophila persimilis RepID=B4G... 53 2e-07
UniRef100_UPI00019859A1 PREDICTED: hypothetical protein n=1 Tax=... 60 2e-07
UniRef100_B4JKF1 GH12063 n=1 Tax=Drosophila grimshawi RepID=B4JK... 57 3e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 61 3e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 61 3e-07
UniRef100_B3Z1U5 Putative uncharacterized protein n=1 Tax=Bacill... 61 3e-07
UniRef100_A3WEW6 Peptidoglycan-associated protein n=1 Tax=Erythr... 61 3e-07
UniRef100_Q2PF07 Putative uncharacterized protein (Fragment) n=1... 61 3e-07
UniRef100_B9PDE3 Predicted protein (Fragment) n=1 Tax=Populus tr... 61 3e-07
UniRef100_A8J8Z8 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 61 3e-07
UniRef100_Q9GRW7 Protein no-on-transient A n=1 Tax=Drosophila vi... 61 3e-07
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 54 3e-07
UniRef100_UPI000194BCD9 PREDICTED: similar to hCG38312 n=1 Tax=T... 60 4e-07
UniRef100_UPI00017466C4 RNA-binding protein (RRM domain) n=1 Tax... 60 4e-07
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 60 4e-07
UniRef100_Q3ANN6 RNA-binding region RNP-1 (RNA recognition motif... 60 4e-07
UniRef100_A3WBS5 Peptidoglycan-associated protein n=1 Tax=Erythr... 60 4e-07
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 60 4e-07
UniRef100_B4JMZ8 GH24721 n=1 Tax=Drosophila grimshawi RepID=B4JM... 60 4e-07
UniRef100_B3MYN1 GF22178 n=1 Tax=Drosophila ananassae RepID=B3MY... 60 4e-07
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 55 4e-07
UniRef100_B4GDZ6 GL21920 n=1 Tax=Drosophila persimilis RepID=B4G... 47 4e-07
UniRef100_Q1A1A3 Forkhead box protein G1 n=1 Tax=Epomophorus gam... 44 4e-07
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 49 5e-07
UniRef100_B4MEV8 GJ14893 n=1 Tax=Drosophila virilis RepID=B4MEV8... 51 5e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 60 5e-07
UniRef100_B9LBU3 Putative uncharacterized protein n=1 Tax=Chloro... 60 5e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 60 5e-07
UniRef100_A9WHI6 Putative uncharacterized protein n=1 Tax=Chloro... 60 5e-07
UniRef100_A1T5K2 Putative uncharacterized protein n=1 Tax=Mycoba... 60 5e-07
UniRef100_B5FWY8 OmpA family protein n=1 Tax=Riemerella anatipes... 60 5e-07
UniRef100_C5XP48 Putative uncharacterized protein Sb03g005056 (F... 60 5e-07
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 60 5e-07
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 60 5e-07
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 60 5e-07
UniRef100_B7PAV3 Putative uncharacterized protein (Fragment) n=1... 60 5e-07
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 60 5e-07
UniRef100_B0CT66 RhoA GTPase effector DIA/Diaphanous n=1 Tax=Lac... 60 5e-07
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 60 5e-07
UniRef100_Q15637-5 Isoform 5 of Splicing factor 1 n=1 Tax=Homo s... 60 5e-07
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 59 5e-07
UniRef100_Q1A1A2 Forkhead box protein G1 n=1 Tax=Equus burchelli... 44 5e-07
UniRef100_UPI0000D9B690 PREDICTED: similar to diaphanous 1 n=1 T... 45 6e-07
UniRef100_Q4P876 Putative uncharacterized protein n=1 Tax=Ustila... 47 6e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 60 6e-07
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 60 6e-07
UniRef100_UPI00005EB969 PREDICTED: similar to SET binding protei... 60 6e-07
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 60 6e-07
UniRef100_UPI00015DF6E5 enabled homolog (Drosophila) n=1 Tax=Mus... 60 6e-07
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 60 6e-07
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 60 6e-07
UniRef100_UPI000179D080 UPI000179D080 related cluster n=1 Tax=Bo... 60 6e-07
UniRef100_Q2N609 Peptidoglycan-associated protein n=1 Tax=Erythr... 60 6e-07
UniRef100_A4TEZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 60 6e-07
UniRef100_Q1RH03 Cell surface antigen Sca2 n=2 Tax=Rickettsia be... 60 6e-07
UniRef100_Q3L8Q3 Surface antigen (Fragment) n=1 Tax=Rickettsia b... 60 6e-07
UniRef100_Q9ZWT0 Extensin n=1 Tax=Adiantum capillus-veneris RepI... 60 6e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 60 6e-07
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 60 6e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 60 6e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 60 6e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 60 6e-07
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 60 6e-07
UniRef100_B4H321 GL13352 n=1 Tax=Drosophila persimilis RepID=B4H... 60 6e-07
UniRef100_A8QF93 EBNA-2 nuclear protein, putative n=1 Tax=Brugia... 60 6e-07
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 60 6e-07
UniRef100_B5XQY2 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 60 6e-07
UniRef100_P04264 Keratin, type II cytoskeletal 1 n=1 Tax=Homo sa... 54 7e-07
UniRef100_B4I386 GM18470 n=1 Tax=Drosophila sechellia RepID=B4I3... 52 7e-07
UniRef100_Q8I1F4 rRNA 2'-O-methyltransferase fibrillarin n=1 Tax... 57 7e-07
UniRef100_C1ECP1 Resistance-nodulation-cell division superfamily... 42 7e-07
UniRef100_Q54WH2 Formin-A n=1 Tax=Dictyostelium discoideum RepID... 47 8e-07
UniRef100_UPI0000ECBAD6 inverted formin 2 isoform 1 n=1 Tax=Gall... 42 8e-07
UniRef100_Q7QC89 AGAP002506-PA (Fragment) n=1 Tax=Anopheles gamb... 42 8e-07
UniRef100_Q39337 Glycine-rich_protein_(Aa1-291) n=1 Tax=Brassica... 53 8e-07
UniRef100_UPI0001552F36 PREDICTED: similar to SH3 domain binding... 59 8e-07
UniRef100_UPI0001552DAA PREDICTED: similar to CR16 n=1 Tax=Mus m... 59 8e-07
UniRef100_UPI0000E491FD PREDICTED: hypothetical protein, partial... 59 8e-07
UniRef100_Q9XIL9 Putative uncharacterized protein At2g15880 n=1 ... 59 8e-07
UniRef100_Q0PIW3 HyPRP1 n=1 Tax=Gossypium hirsutum RepID=Q0PIW3_... 59 8e-07
UniRef100_C1N0Z7 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 8e-07
UniRef100_Q5CS67 Signal peptide containing large protein with pr... 59 8e-07
UniRef100_Q7YY78 Protease, possible n=2 Tax=Cryptosporidium parv... 59 8e-07
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 59 8e-07
UniRef100_B4IF47 GM13418 n=1 Tax=Drosophila sechellia RepID=B4IF... 59 8e-07
UniRef100_A7UTZ4 AGAP005965-PA (Fragment) n=1 Tax=Anopheles gamb... 59 8e-07
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 59 8e-07
UniRef100_A2R4H0 Function: sepA function in A. nidulans requires... 59 8e-07
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 59 8e-07
UniRef100_P0C7L0-2 Isoform 2 of WAS/WASL-interacting protein fam... 59 8e-07
UniRef100_P0C7L0 WAS/WASL-interacting protein family member 3 n=... 59 8e-07
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 59 8e-07
UniRef100_UPI0000F3293E PREDICTED: Bos taurus similar to bromodo... 45 9e-07
UniRef100_Q4TB22 Chromosome 15 SCAF7210, whole genome shotgun se... 55 9e-07
UniRef100_A6RGJ8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 55 9e-07
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 58 9e-07
UniRef100_Q1A1A6 Forkhead box protein G1 n=1 Tax=Cebus capucinus... 44 9e-07
UniRef100_Q39835 Extensin n=1 Tax=Glycine max RepID=Q39835_SOYBN 52 9e-07
UniRef100_Q9C946 Putative uncharacterized protein T7P1.21 n=1 Ta... 42 1e-06
UniRef100_C5YH07 Putative uncharacterized protein Sb07g003820 n=... 50 1e-06
UniRef100_Q7Y218 Putative uncharacterized protein At5g46730 n=1 ... 53 1e-06
UniRef100_Q9FIQ2 Genomic DNA, chromosome 5, P1 clone:MZA15 n=1 T... 53 1e-06
UniRef100_B3H4R1 Uncharacterized protein At5g46730.2 n=1 Tax=Ara... 53 1e-06
UniRef100_UPI0001B58635 hypothetical protein StAA4_25414 n=1 Tax... 59 1e-06
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 59 1e-06
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 59 1e-06
UniRef100_UPI00016E7FC0 UPI00016E7FC0 related cluster n=1 Tax=Ta... 59 1e-06
UniRef100_B0SVF7 OmpA/MotB domain protein n=1 Tax=Caulobacter sp... 59 1e-06
UniRef100_A5GPV8 RNA-binding protein, RRM domain n=1 Tax=Synecho... 59 1e-06
UniRef100_A1UCC3 Putative uncharacterized protein n=2 Tax=Mycoba... 59 1e-06
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 59 1e-06
UniRef100_C5XW81 Putative uncharacterized protein Sb04g005115 n=... 59 1e-06
UniRef100_B9RS81 Serine-threonine protein kinase, plant-type, pu... 59 1e-06
UniRef100_Q5CLH8 Protease n=1 Tax=Cryptosporidium hominis RepID=... 59 1e-06
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 59 1e-06
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 59 1e-06
UniRef100_Q84W21 Putative uncharacterized protein At1g62240 n=1 ... 48 1e-06
UniRef100_Q388E8 ATP-dependent DEAD/H RNA helicase, putative n=1... 55 1e-06
UniRef100_D0A0A9 ATP-dependent DEAD/H RNA helicase, putative n=1... 55 1e-06
UniRef100_Q8LPA7 Cold shock protein-1 n=1 Tax=Triticum aestivum ... 55 1e-06
UniRef100_B4M8D6 GJ16790 n=1 Tax=Drosophila virilis RepID=B4M8D6... 49 1e-06
UniRef100_B4L7Q3 GI11213 n=1 Tax=Drosophila mojavensis RepID=B4L... 50 1e-06
UniRef100_UPI00017603E5 PREDICTED: diaphanous 2 n=1 Tax=Danio re... 59 1e-06
UniRef100_UPI0000F2E99A PREDICTED: similar to diaphanous homolog... 59 1e-06
UniRef100_UPI0000E468D6 PREDICTED: similar to laminin alpha chai... 59 1e-06
UniRef100_UPI0000E23E7D PREDICTED: WAS protein homology region 2... 59 1e-06
UniRef100_UPI0000603C6F PREDICTED: Ras association (RalGDS/AF-6)... 59 1e-06
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 59 1e-06
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 59 1e-06
UniRef100_B4U926 Putative uncharacterized protein n=1 Tax=Hydrog... 59 1e-06
UniRef100_A3Q1Z8 Molecular chaperone-like n=3 Tax=Mycobacterium ... 59 1e-06
UniRef100_B3Z1W8 Putative uncharacterized protein (Fragment) n=1... 59 1e-06
UniRef100_A3WBR8 Peptidoglycan-associated protein n=1 Tax=Erythr... 59 1e-06
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 59 1e-06
UniRef100_Q6EUQ1 Os02g0176100 protein n=1 Tax=Oryza sativa Japon... 59 1e-06
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 59 1e-06
UniRef100_B9H7Q5 Predicted protein n=1 Tax=Populus trichocarpa R... 59 1e-06
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 59 1e-06
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 59 1e-06
UniRef100_B7Q5W9 Secreted protein, putative (Fragment) n=1 Tax=I... 59 1e-06
UniRef100_B7PNV6 Putative uncharacterized protein (Fragment) n=1... 59 1e-06
UniRef100_B4R5R3 GD17265 n=1 Tax=Drosophila simulans RepID=B4R5R... 59 1e-06
UniRef100_B4PVU3 GE23484 n=1 Tax=Drosophila yakuba RepID=B4PVU3_... 59 1e-06
UniRef100_B3RJQ3 Putative uncharacterized protein n=1 Tax=Tricho... 59 1e-06
UniRef100_C9K0J5 Putative uncharacterized protein RAPH1 n=1 Tax=... 59 1e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 59 1e-06
UniRef100_Q70E73 Ras-associated and pleckstrin homology domains-... 59 1e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 59 1e-06
UniRef100_UPI000180CA59 PREDICTED: similar to formin, inverted n... 52 1e-06
UniRef100_UPI0000D9CC90 PREDICTED: similar to keratin 1 isoform ... 52 2e-06
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 55 2e-06
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 52 2e-06
UniRef100_A2XPC7 Putative uncharacterized protein n=1 Tax=Oryza ... 49 2e-06
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 52 2e-06
UniRef100_B3MW45 GF22329 n=1 Tax=Drosophila ananassae RepID=B3MW... 53 2e-06
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 58 2e-06
UniRef100_UPI0000E22B4E PREDICTED: splicing factor 1 isoform 1 n... 58 2e-06
UniRef100_UPI0001B7BC6C UPI0001B7BC6C related cluster n=1 Tax=Ra... 58 2e-06
UniRef100_UPI0000ECC7C5 enabled homolog n=1 Tax=Gallus gallus Re... 58 2e-06
UniRef100_UPI0000ECC7C4 enabled homolog n=1 Tax=Gallus gallus Re... 58 2e-06
UniRef100_UPI0000ECC7C3 enabled homolog n=1 Tax=Gallus gallus Re... 58 2e-06
UniRef100_UPI00003ACBF3 enabled homolog n=1 Tax=Gallus gallus Re... 58 2e-06
UniRef100_Q9DEG2 AvenaII n=1 Tax=Gallus gallus RepID=Q9DEG2_CHICK 58 2e-06
UniRef100_Q9DEG1 AvenaIII n=1 Tax=Gallus gallus RepID=Q9DEG1_CHICK 58 2e-06
UniRef100_Q90YB5 AvEna neural variant n=1 Tax=Gallus gallus RepI... 58 2e-06
UniRef100_Q0IHK2 Sfrs1 protein (Fragment) n=1 Tax=Xenopus laevis... 58 2e-06
UniRef100_O93263 Avena n=1 Tax=Gallus gallus RepID=O93263_CHICK 58 2e-06
UniRef100_A1A628 Sfrs1 protein (Fragment) n=2 Tax=Xenopus laevis... 58 2e-06
UniRef100_Q1BU84 Putative uncharacterized protein n=1 Tax=Burkho... 58 2e-06
UniRef100_Q0C0P5 OmpA family protein n=1 Tax=Hyphomonas neptuniu... 58 2e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 58 2e-06
UniRef100_A5V8M1 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 58 2e-06
UniRef100_A5V4K4 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 58 2e-06
UniRef100_A0K9V3 Putative uncharacterized protein n=1 Tax=Burkho... 58 2e-06
UniRef100_C6WKJ2 NLP/P60 protein n=1 Tax=Actinosynnema mirum DSM... 58 2e-06
UniRef100_B3Z1W9 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B3Z1W3 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B3Z1V9 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B3Z1V7 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1U9 Ferrichrome-binding protein (Fragment) n=1 Tax=... 58 2e-06
UniRef100_B3Z1U4 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B3Z1U3 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1U1 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1T8 Endopeptidase lactocepin n=1 Tax=Bacillus cereu... 58 2e-06
UniRef100_B3Z1T7 Cell-division initiation protein DivIB n=1 Tax=... 58 2e-06
UniRef100_B3Z1T6 Amino acid permease n=1 Tax=Bacillus cereus W R... 58 2e-06
UniRef100_B3Z1T5 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1T1 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1S9 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_B3Z1S7 Putative uncharacterized protein n=1 Tax=Bacill... 58 2e-06
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 58 2e-06
UniRef100_C1E9S3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 58 2e-06
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 58 2e-06
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 58 2e-06
UniRef100_Q9VD49 CG5778, isoform A n=1 Tax=Drosophila melanogast... 58 2e-06
UniRef100_C1IS34 Minicollagen-1 n=1 Tax=Malo kingi RepID=C1IS34_... 58 2e-06
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B7PJF9 Menin, putative n=1 Tax=Ixodes scapularis RepID... 58 2e-06
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 58 2e-06
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 58 2e-06
UniRef100_B5DH56 GA25354 n=1 Tax=Drosophila pseudoobscura pseudo... 58 2e-06
UniRef100_B4QT65 GD21099 n=1 Tax=Drosophila simulans RepID=B4QT6... 58 2e-06
UniRef100_B2AKS1 Translation initiation factor IF-2 n=1 Tax=Podo... 58 2e-06
UniRef100_A7EUH8 Putative uncharacterized protein n=1 Tax=Sclero... 58 2e-06
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 58 2e-06
UniRef100_O60885 Bromodomain-containing protein 4 n=1 Tax=Homo s... 44 2e-06
UniRef100_UPI0000E25017 PREDICTED: bromodomain-containing protei... 44 2e-06
UniRef100_Q9LSN6 Genomic DNA, chromosome 3, TAC clone:K14A17 n=1... 50 2e-06
UniRef100_UPI0000D9CC8C PREDICTED: similar to keratin 1 isoform ... 52 2e-06
UniRef100_UPI0000D9CC8D PREDICTED: similar to keratin 1 isoform ... 52 2e-06
UniRef100_UPI0000D9CC8E PREDICTED: similar to keratin 1 isoform ... 52 2e-06
UniRef100_Q29LJ4 GA17232 n=1 Tax=Drosophila pseudoobscura pseudo... 53 2e-06
UniRef100_UPI0000D9CC8F PREDICTED: similar to keratin 1 isoform ... 52 2e-06
UniRef100_B8A7W6 Putative uncharacterized protein n=1 Tax=Oryza ... 54 2e-06
UniRef100_UPI0000EBE04E PREDICTED: similar to brain factor-1 iso... 44 2e-06
UniRef100_A2YER4 Putative uncharacterized protein n=1 Tax=Oryza ... 57 2e-06
UniRef100_A0E1N2 Chromosome undetermined scaffold_73, whole geno... 56 2e-06
UniRef100_UPI0000F22463 IQ motif and Sec7 domain 2 n=1 Tax=Mus m... 42 2e-06
UniRef100_Q5JU85-2 Isoform 2 of IQ motif and SEC7 domain-contain... 42 2e-06
UniRef100_A4GZ26 ARF6 guanine nucleotide exchange factor IQArfGE... 42 2e-06
UniRef100_UPI00015DF451 IQ motif and Sec7 domain 2 n=1 Tax=Mus m... 42 2e-06
UniRef100_Q5DU25 IQ motif and SEC7 domain-containing protein 2 n... 42 2e-06
UniRef100_Q5JU85 IQ motif and SEC7 domain-containing protein 2 n... 42 2e-06
UniRef100_UPI000179E2B1 UPI000179E2B1 related cluster n=1 Tax=Bo... 42 2e-06
UniRef100_Q39864 Hydroxyproline-rich glycoprotein (Fragment) n=1... 50 2e-06
UniRef100_UPI0001797DF3 PREDICTED: similar to IQ motif and Sec7 ... 42 2e-06
UniRef100_B9RFX3 DNA binding protein, putative n=1 Tax=Ricinus c... 52 2e-06
UniRef100_UPI0001553749 PREDICTED: hypothetical protein n=1 Tax=... 58 2e-06
UniRef100_C6XK60 OmpA/MotB domain protein n=1 Tax=Hirschia balti... 58 2e-06
UniRef100_B0T768 TonB family protein n=1 Tax=Caulobacter sp. K31... 58 2e-06
UniRef100_A3PW04 U5 snRNP spliceosome subunit-like protein n=1 T... 58 2e-06
UniRef100_A4CWS8 RNA-binding region RNP-1 (RNA recognition motif... 58 2e-06
UniRef100_C0PGT8 Putative uncharacterized protein n=1 Tax=Zea ma... 58 2e-06
UniRef100_A8JAG8 Argonaute-like protein n=1 Tax=Chlamydomonas re... 58 2e-06
UniRef100_Q9GP44 NONA protein (Fragment) n=1 Tax=Drosophila viri... 58 2e-06
UniRef100_Q95UX4 No on or off transient A (Fragment) n=1 Tax=Dro... 58 2e-06
UniRef100_Q95NR6 No on or off transient A (Fragment) n=1 Tax=Dro... 58 2e-06
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 58 2e-06
UniRef100_B4NC76 GK25807 n=1 Tax=Drosophila willistoni RepID=B4N... 58 2e-06
UniRef100_B4MID6 GK23346 n=1 Tax=Drosophila willistoni RepID=B4M... 58 2e-06
UniRef100_B4KSC4 GI19596 n=1 Tax=Drosophila mojavensis RepID=B4K... 58 2e-06
UniRef100_B3N1Q5 GF20750 n=1 Tax=Drosophila ananassae RepID=B3N1... 58 2e-06
UniRef100_B3MV25 GF22854 n=1 Tax=Drosophila ananassae RepID=B3MV... 58 2e-06
UniRef100_B0WEX8 Putative uncharacterized protein n=1 Tax=Culex ... 58 2e-06
UniRef100_C6HGV1 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 58 2e-06
UniRef100_C0NZQ8 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 58 2e-06
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 58 2e-06
UniRef100_A0DA74 Chromosome undetermined scaffold_43, whole geno... 40 3e-06
UniRef100_UPI0000E25D49 PREDICTED: similar to IQ motif and Sec7 ... 42 3e-06
UniRef100_B4J560 GH20878 n=1 Tax=Drosophila grimshawi RepID=B4J5... 41 3e-06
UniRef100_C5YQE0 Putative uncharacterized protein Sb08g019190 n=... 48 3e-06
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 53 3e-06
UniRef100_UPI000184A164 Bromodomain-containing protein 4 (HUNK1 ... 43 3e-06
UniRef100_UPI000194BDE1 PREDICTED: zinc finger homeodomain 4 n=1... 57 3e-06
UniRef100_UPI0001795F40 PREDICTED: leiomodin 2 (cardiac) n=1 Tax... 57 3e-06
UniRef100_UPI000151AF18 hypothetical protein PGUG_03629 n=1 Tax=... 57 3e-06
UniRef100_UPI0000F2E472 PREDICTED: hypothetical protein n=1 Tax=... 57 3e-06
UniRef100_UPI0000DA1CBA PREDICTED: hypothetical protein n=1 Tax=... 57 3e-06
UniRef100_UPI0000D9E82E PREDICTED: similar to additional sex com... 57 3e-06
UniRef100_UPI0000D9E82D PREDICTED: similar to additional sex com... 57 3e-06
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 57 3e-06
UniRef100_B6IMJ6 Putative uncharacterized protein n=1 Tax=Rhodos... 57 3e-06
UniRef100_A2C5L0 RNA-binding region RNP-1 (RNA recognition motif... 57 3e-06
UniRef100_Q062H8 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 57 3e-06
UniRef100_A5P9Z0 Peptidoglycan-associated protein n=1 Tax=Erythr... 57 3e-06
UniRef100_Q3ECQ3 Uncharacterized protein At1g54215.1 n=1 Tax=Ara... 57 3e-06
UniRef100_Q39599 Extensin n=1 Tax=Catharanthus roseus RepID=Q395... 57 3e-06
UniRef100_C5YU09 Putative uncharacterized protein Sb08g008380 n=... 57 3e-06
UniRef100_A9T0V1 Predicted protein n=1 Tax=Physcomitrella patens... 57 3e-06
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 57 3e-06
UniRef100_C0MP97 Formin-homology protein SmDia n=1 Tax=Schistoso... 57 3e-06
UniRef100_B7P671 Cement protein, putative n=1 Tax=Ixodes scapula... 57 3e-06
UniRef100_B4M1S7 NonA n=1 Tax=Drosophila virilis RepID=B4M1S7_DROVI 57 3e-06
UniRef100_B4L8T7 GI14292 n=1 Tax=Drosophila mojavensis RepID=B4L... 57 3e-06
UniRef100_B3NX66 GG17948 n=1 Tax=Drosophila erecta RepID=B3NX66_... 57 3e-06
UniRef100_A8NTE1 Formin Homology 2 Domain containing protein n=1... 57 3e-06
UniRef100_A8NM82 Formin Homology 2 Domain containing protein (Fr... 57 3e-06
UniRef100_B7Z9A8 cDNA FLJ58392 n=1 Tax=Homo sapiens RepID=B7Z9A8... 57 3e-06
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 57 3e-06
UniRef100_A8NHF1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 57 3e-06
UniRef100_Q1A1A4 Forkhead box protein G1 n=1 Tax=Pipistrellus ru... 41 3e-06
UniRef100_C9J285 Putative uncharacterized protein ENSP0000039240... 54 3e-06
UniRef100_B4QI53 GD25085 n=1 Tax=Drosophila simulans RepID=B4QI5... 55 3e-06
UniRef100_A4R4Z1 Putative uncharacterized protein n=1 Tax=Magnap... 50 4e-06
UniRef100_UPI0001926BAB PREDICTED: similar to mini-collagen n=1 ... 57 4e-06
UniRef100_UPI0000F2BFBF PREDICTED: similar to bromodomain PHD fi... 57 4e-06
UniRef100_UPI0000DB6FAC PREDICTED: similar to Protein cappuccino... 57 4e-06
UniRef100_UPI0000DA313E PREDICTED: similar to CG11835-PA n=1 Tax... 57 4e-06
UniRef100_UPI00005E7A0E PREDICTED: similar to HOXD8 n=1 Tax=Mono... 57 4e-06
UniRef100_UPI00005A3937 PREDICTED: similar to formin-like 2 isof... 57 4e-06
UniRef100_UPI0001B7A7A5 UPI0001B7A7A5 related cluster n=1 Tax=Ra... 57 4e-06
UniRef100_UPI0001548235 nucleolar protein family A, member 1 (H/... 57 4e-06
UniRef100_B1KJL7 Putative lipoprotein n=1 Tax=Shewanella woodyi ... 57 4e-06
UniRef100_A9FZR5 Single-stranded DNA-binding protein n=1 Tax=Sor... 57 4e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 57 4e-06
UniRef100_Q39600 Extensin n=1 Tax=Catharanthus roseus RepID=Q396... 57 4e-06
UniRef100_Q00ZZ2 Membrane coat complex Retromer, subunit VPS5/SN... 57 4e-06
UniRef100_B9H173 Predicted protein n=1 Tax=Populus trichocarpa R... 57 4e-06
UniRef100_B8B0N9 Putative uncharacterized protein n=1 Tax=Oryza ... 57 4e-06
UniRef100_A9RSY7 Predicted protein n=1 Tax=Physcomitrella patens... 57 4e-06
UniRef100_A7YFB9 Glycine-rich protein n=1 Tax=Lilium formosanum ... 57 4e-06
UniRef100_A4RZU2 Predicted protein n=1 Tax=Ostreococcus lucimari... 57 4e-06
UniRef100_C5KAT0 Putative uncharacterized protein n=1 Tax=Perkin... 57 4e-06
UniRef100_C3Z6D8 Putative uncharacterized protein n=1 Tax=Branch... 57 4e-06
UniRef100_B4R5S1 GD17272 n=1 Tax=Drosophila simulans RepID=B4R5S... 57 4e-06
UniRef100_B4L246 GI15365 n=1 Tax=Drosophila mojavensis RepID=B4L... 57 4e-06
UniRef100_B4HH21 GM26598 n=1 Tax=Drosophila sechellia RepID=B4HH... 57 4e-06
UniRef100_A8P9A8 Putative uncharacterized protein n=1 Tax=Brugia... 57 4e-06
UniRef100_A4H5Q3 Cleavage and polyadenylation specificity factor... 57 4e-06
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 57 4e-06
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 57 4e-06
UniRef100_A5DK28 Putative uncharacterized protein n=1 Tax=Pichia... 57 4e-06
UniRef100_A3AB67 Formin-like protein 16 n=1 Tax=Oryza sativa Jap... 57 4e-06
UniRef100_Q07202 Cold and drought-regulated protein CORA n=1 Tax... 57 4e-06
UniRef100_Q91Z58 Uncharacterized protein C6orf132 homolog n=1 Ta... 57 4e-06
UniRef100_UPI0000F2C114 PREDICTED: similar to PR domain containi... 40 4e-06
UniRef100_Q9W1V3 rRNA 2'-O-methyltransferase fibrillarin n=1 Tax... 54 4e-06
UniRef100_B3MF29 GF11304 n=1 Tax=Drosophila ananassae RepID=B3MF... 53 4e-06
UniRef100_Q9M6R7 Extensin n=1 Tax=Pisum sativum RepID=Q9M6R7_PEA 51 4e-06
UniRef100_B3NK90 GG20960 n=1 Tax=Drosophila erecta RepID=B3NK90_... 45 4e-06
UniRef100_Q02021 Glycine-rich protein n=2 Tax=Solanum lycopersic... 50 5e-06