Tomato SBM (selected BAC clone mixture) contigs
To enrich gene-rich regions within the tomato genome, two sets of BAC clone pools were generated using the BAC end sequences available at the SGN website, and denoted as SBM (selected BAC clone mixture).
In the first set (SBM-I), a total of 20,000 BAC clones, in which the sequence at neither end showed any homology to any of the tomato repetitive sequences registered in the SGN tomato genome repeat data, were selected from the BAC libraries (10,000 clones from the HindIII library, and 5,000 clones each from the MboI and EcoRI libraries).
The second set (SBM-II) selected included clones that contained repetitive sequences at one end but unique sequences at the other. A total of 10,800 clones (5,400 clones from the HindIII library, and 2,700 clones each from the MboI and EcoRI libraries) were pooled.
Shotgun libraries were generated from the SBM-I and SBM-II sets, and sequencing was performed by the Sanger method. The total number of accumulated reads was 4,248,000: 2,916,000 from SBM-I and 1,332,000 from SBM-II. The sequences were then subjected to assembly, along with the BAC and fosmid end sequences that had been made publicly available by SGN. As a result, non-redundant sequences of 540,588,968 bp that consisted of 100,783 contigs have been generated. The statistics of the assembly are summarized here.
Scaffolds were formed by ordering and orienting the contig sequences, by taking the relative positions of the BAC and fosmid end sequences into account. The number of scaffolds thus generated was 3,996 containing 15,119 contigs.
The sequence information of the SBM contigs is available through the public DNA databases (DDBJ/Genbank/EMBL) under the accession numbers BABP01000001-BABP01100783.
Contig name
Assembled contigs, excluding those shorter than 1,000 bp in size, were designated as SlSBM (Solanum lycopersicum Selected BAC clone Mixture) followed by the assembly consensus names containing an indication of the assembly status. Note that these contig names should be regarded as temporal. The SBM sequence data are currently being integrated into the whole genome assembly data by the International Tomato Sequencing Consortium, and new standardized contig names will be provided in the future.
-
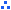 SlSBM_S*****_**
SlSBM_S*****_**
The contigs in the scaffold groups are designated by the alphabetical code "S" and a five-digit group number followed by a two-digit sequential number for each contig.
-
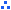 SlSBM_L*****_01
SlSBM_L*****_01
The singlet contigs containing the BAC and fosmid-end sequences as assembly components are indicated by the alphabetical code "L" and a five-digit contig number followed by the consensus code "01".
-
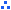 SlSBM_*****_01
SlSBM_*****_01
The contigs exclusively composed of SlSBM shotgun sequences are designated with a six-digit contig number followed by the consensus code 01.