[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= DC597777 MPDL024d04_r
(335 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
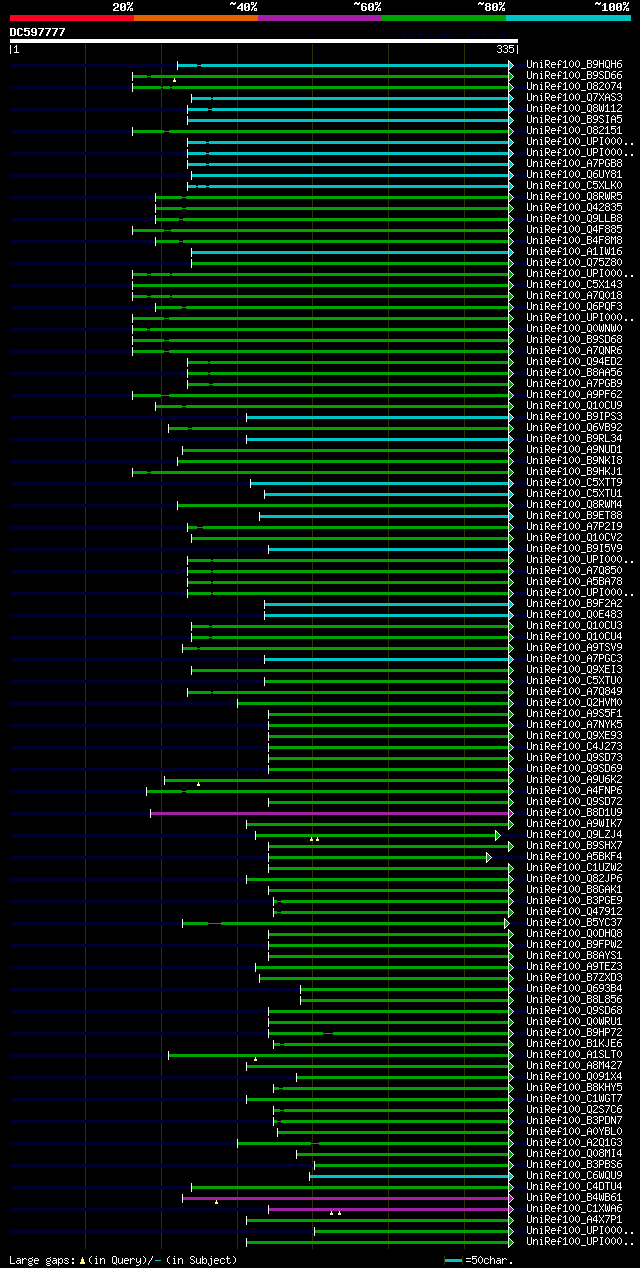
significant alignments:[graphical|details]
UniRef100_B9HQH6 Predicted protein n=1 Tax=Populus trichocarpa R... 111 3e-23
UniRef100_B9SD66 Hydrolase, hydrolyzing O-glycosyl compounds, pu... 109 8e-23
UniRef100_O82074 Beta-D-glucosidase n=1 Tax=Tropaeolum majus Rep... 108 1e-22
UniRef100_Q7XAS3 Beta-D-glucosidase n=1 Tax=Gossypium hirsutum R... 106 9e-22
UniRef100_Q8W112 Beta-D-glucan exohydrolase-like protein n=1 Tax... 101 2e-20
UniRef100_B9SIA5 Hydrolase, hydrolyzing O-glycosyl compounds, pu... 101 2e-20
UniRef100_O82151 Beta-D-glucan exohydrolase n=1 Tax=Nicotiana ta... 101 3e-20
UniRef100_UPI00019837C8 PREDICTED: hypothetical protein isoform ... 98 2e-19
UniRef100_UPI00019837C7 PREDICTED: hypothetical protein isoform ... 98 2e-19
UniRef100_A7PGB8 Chromosome chr6 scaffold_15, whole genome shotg... 98 2e-19
UniRef100_Q6UY81 Exo-beta-glucanase n=1 Tax=Lilium longiflorum R... 98 3e-19
UniRef100_C5XLK0 Putative uncharacterized protein Sb03g035970 n=... 97 6e-19
UniRef100_Q8RWR5 Beta-D-glucan exohydrolase n=1 Tax=Triticum aes... 96 1e-18
UniRef100_Q42835 Beta-D-glucan exohydrolase, isoenzyme ExoII n=1... 96 1e-18
UniRef100_Q9LLB8 Exoglucanase n=1 Tax=Zea mays RepID=Q9LLB8_MAIZE 96 2e-18
UniRef100_Q4F885 Endo-alpha-1,4-glucanase n=1 Tax=Gossypium hirs... 96 2e-18
UniRef100_B4F8M8 Putative uncharacterized protein n=1 Tax=Zea ma... 96 2e-18
UniRef100_A1IW16 Beta-1,3-glucanase (PR protein) (Fragment) n=1 ... 96 2e-18
UniRef100_Q75Z80 Exo-1,3-beta-glucanase n=1 Tax=Lilium longiflor... 95 3e-18
UniRef100_UPI0001983BFB PREDICTED: hypothetical protein n=1 Tax=... 94 4e-18
UniRef100_C5X143 Putative uncharacterized protein Sb01g008050 n=... 94 4e-18
UniRef100_A7Q018 Chromosome chr8 scaffold_41, whole genome shotg... 94 4e-18
UniRef100_Q6PQF3 Cell wall beta-glucosidase n=1 Tax=Secale cerea... 93 8e-18
UniRef100_UPI0001985AB6 PREDICTED: hypothetical protein n=1 Tax=... 93 1e-17
UniRef100_Q0WNW0 Putative uncharacterized protein At5g04885 n=2 ... 93 1e-17
UniRef100_B9SD68 Hydrolase, hydrolyzing O-glycosyl compounds, pu... 93 1e-17
UniRef100_A7QNR6 Chromosome undetermined scaffold_134, whole gen... 93 1e-17
UniRef100_Q94ED2 Os01g0771900 protein n=1 Tax=Oryza sativa Japon... 91 3e-17
UniRef100_B8AA56 Putative uncharacterized protein n=1 Tax=Oryza ... 91 3e-17
UniRef100_A7PGB9 Chromosome chr6 scaffold_15, whole genome shotg... 91 4e-17
UniRef100_A9PF62 Putative uncharacterized protein n=1 Tax=Populu... 89 1e-16
UniRef100_Q10CU9 Os03g0749300 protein n=3 Tax=Oryza sativa RepID... 89 1e-16
UniRef100_B9IPS3 Predicted protein n=1 Tax=Populus trichocarpa R... 89 2e-16
UniRef100_Q6VB92 Beta-glucanase n=1 Tax=Zea mays RepID=Q6VB92_MAIZE 89 2e-16
UniRef100_B9RL34 Periplasmic beta-glucosidase, putative n=1 Tax=... 89 2e-16
UniRef100_A9NUD1 Putative uncharacterized protein n=1 Tax=Picea ... 89 2e-16
UniRef100_B9NKI8 Predicted protein (Fragment) n=1 Tax=Populus tr... 88 3e-16
UniRef100_B9HKJ1 Predicted protein n=1 Tax=Populus trichocarpa R... 87 4e-16
UniRef100_C5XTT9 Putative uncharacterized protein Sb04g002560 n=... 87 6e-16
UniRef100_C5XTU1 Putative uncharacterized protein Sb04g002580 n=... 86 1e-15
UniRef100_Q8RWM4 Beta-glucosidase-like protein n=1 Tax=Arabidops... 86 2e-15
UniRef100_B9ET88 Putative uncharacterized protein n=1 Tax=Oryza ... 85 3e-15
UniRef100_A7P2I9 Chromosome chr1 scaffold_5, whole genome shotgu... 84 4e-15
UniRef100_Q10CV2 Os03g0749100 protein n=4 Tax=Oryza sativa RepID... 84 5e-15
UniRef100_B9I5V9 Predicted protein n=1 Tax=Populus trichocarpa R... 84 6e-15
UniRef100_UPI0001985AE8 PREDICTED: hypothetical protein n=1 Tax=... 83 8e-15
UniRef100_A7Q850 Chromosome undetermined scaffold_62, whole geno... 83 8e-15
UniRef100_A5BA78 Putative uncharacterized protein n=1 Tax=Vitis ... 83 8e-15
UniRef100_UPI0001985AE7 PREDICTED: hypothetical protein n=1 Tax=... 82 1e-14
UniRef100_B9F2A2 Putative uncharacterized protein n=1 Tax=Oryza ... 80 5e-14
UniRef100_Q0E483 Os02g0131400 protein n=3 Tax=Oryza sativa RepID... 80 5e-14
UniRef100_Q10CU3 Glycosyl hydrolase family 3 N terminal domain c... 79 2e-13
UniRef100_Q10CU4 Os03g0749500 protein n=4 Tax=Oryza sativa RepID... 79 2e-13
UniRef100_A9TSV9 Predicted protein n=1 Tax=Physcomitrella patens... 78 3e-13
UniRef100_A7PGC3 Chromosome chr6 scaffold_15, whole genome shotg... 77 6e-13
UniRef100_Q9XEI3 Beta-D-glucan exohydrolase isoenzyme ExoI n=1 T... 77 8e-13
UniRef100_C5XTU0 Putative uncharacterized protein Sb04g002570 n=... 75 3e-12
UniRef100_A7Q849 Chromosome undetermined scaffold_62, whole geno... 75 3e-12
UniRef100_Q2HVM0 Glycoside hydrolase, family 3, N-terminal; Glyc... 74 5e-12
UniRef100_A9S5F1 Predicted protein n=1 Tax=Physcomitrella patens... 73 9e-12
UniRef100_A7NYK5 Chromosome chr6 scaffold_3, whole genome shotgu... 72 1e-11
UniRef100_Q9XE93 Exhydrolase II n=1 Tax=Zea mays RepID=Q9XE93_MAIZE 70 7e-11
UniRef100_C4J273 Putative uncharacterized protein n=1 Tax=Zea ma... 70 7e-11
UniRef100_Q9SD73 Beta-D-glucan exohydrolase-like protein n=1 Tax... 69 2e-10
UniRef100_Q9SD69 Beta-D-glucan exohydrolase-like protein n=1 Tax... 67 5e-10
UniRef100_A9U6K2 Predicted protein n=1 Tax=Physcomitrella patens... 67 6e-10
UniRef100_A4FNP6 Glucan 1,4-beta-glucosidase n=1 Tax=Saccharopol... 67 8e-10
UniRef100_Q9SD72 Beta-D-glucan exohydrolase-like protein n=2 Tax... 67 8e-10
UniRef100_B8D1U9 Beta-N-acetylhexosaminidase n=1 Tax=Halothermot... 66 1e-09
UniRef100_A9WIK7 Glycoside hydrolase family 3 domain protein n=2... 66 1e-09
UniRef100_Q9LZJ4 Beta-D-glucan exohydrolase-like protein n=1 Tax... 65 2e-09
UniRef100_B9SHX7 Hydrolase, hydrolyzing O-glycosyl compounds, pu... 64 5e-09
UniRef100_A5BKF4 Putative uncharacterized protein n=1 Tax=Vitis ... 64 7e-09
UniRef100_C1UZW2 Beta-glucosidase-like glycosyl hydrolase n=1 Ta... 62 2e-08
UniRef100_Q82JP6 Putative glycosyl hydrolase n=1 Tax=Streptomyce... 61 4e-08
UniRef100_B8GAK1 Glycoside hydrolase family 3 domain protein n=1... 61 4e-08
UniRef100_B3PGE9 Glucan 1,4-beta-glucosidase cel3A n=1 Tax=Cellv... 60 8e-08
UniRef100_Q47912 1,4-B-D-glucan glucohydrolase n=1 Tax=Cellvibri... 60 8e-08
UniRef100_B5YC37 Beta-D-glucosidase n=1 Tax=Dictyoglomus thermop... 60 1e-07
UniRef100_Q0DHQ8 Os05g0449600 protein (Fragment) n=2 Tax=Oryza s... 60 1e-07
UniRef100_B9FPW2 Putative uncharacterized protein n=1 Tax=Oryza ... 60 1e-07
UniRef100_B8AYS1 Putative uncharacterized protein n=1 Tax=Oryza ... 60 1e-07
UniRef100_A9TEZ3 Predicted protein n=1 Tax=Physcomitrella patens... 60 1e-07
UniRef100_B7ZXD3 Putative uncharacterized protein n=1 Tax=Zea ma... 59 2e-07
UniRef100_Q693B4 1,4-beta-D-glucan glucohydrolase n=1 Tax=Microb... 59 2e-07
UniRef100_B8L856 1,4-beta-D-glucan glucohydrolase D n=1 Tax=Sten... 59 2e-07
UniRef100_Q9SD68 Beta-D-glucan exohydrolase-like protein n=1 Tax... 59 2e-07
UniRef100_Q0WRU1 Beta-D-glucan exohydrolase-like protein (Fragme... 59 2e-07
UniRef100_B9HP72 Predicted protein n=1 Tax=Populus trichocarpa R... 59 2e-07
UniRef100_B1KJE6 Glycoside hydrolase family 3 domain protein n=1... 58 4e-07
UniRef100_A1SLT0 Beta-glucosidase n=1 Tax=Nocardioides sp. JS614... 58 4e-07
UniRef100_A8M427 Glycoside hydrolase family 3 domain protein n=1... 57 5e-07
UniRef100_Q091X4 1,4-beta-D-glucan glucohydrolase D n=1 Tax=Stig... 57 6e-07
UniRef100_B8KHY5 Glucan 1,4-beta-glucosidase n=1 Tax=gamma prote... 57 6e-07
UniRef100_C1WGT7 Beta-glucosidase-like glycosyl hydrolase n=1 Ta... 57 8e-07
UniRef100_Q2S7C6 Beta-glucosidase-related Glycosidase n=1 Tax=Ha... 56 1e-06
UniRef100_B3PDN7 Glucan 1,4-beta-glucosidase, putative, cel3B n=... 56 1e-06
UniRef100_A0YBL0 Beta-glucosidase-related Glycosidase n=1 Tax=ma... 56 1e-06
UniRef100_A2Q1G3 Glycoside hydrolase, family 3, N-terminal n=1 T... 56 1e-06
UniRef100_Q08MI4 1,4-beta-D-glucan glucohydrolase (Fragment) n=1... 55 2e-06
UniRef100_B3PBS6 Putative 1,4-beta-D-glucan glucohydrolase cel3D... 55 3e-06
UniRef100_C6WQU9 Glycoside hydrolase family 3 domain protein n=1... 55 3e-06
UniRef100_C4DTU4 Beta-glucosidase-like glycosyl hydrolase n=1 Ta... 55 3e-06
UniRef100_B4WB61 Glycosyl hydrolase family 3 N terminal domain p... 55 3e-06
UniRef100_C1XWA6 Beta-glucosidase-like glycosyl hydrolase n=1 Ta... 54 4e-06
UniRef100_A4X7P1 Glycoside hydrolase, family 3 domain protein n=... 54 5e-06
UniRef100_UPI0001B55B29 glycoside hydrolase family 3 domain prot... 54 7e-06
UniRef100_UPI0001AEE95A glycosyl hydrolase n=1 Tax=Streptomyces ... 54 7e-06
UniRef100_B7RZU7 Glycosyl hydrolase family 3 N terminal domain p... 53 9e-06