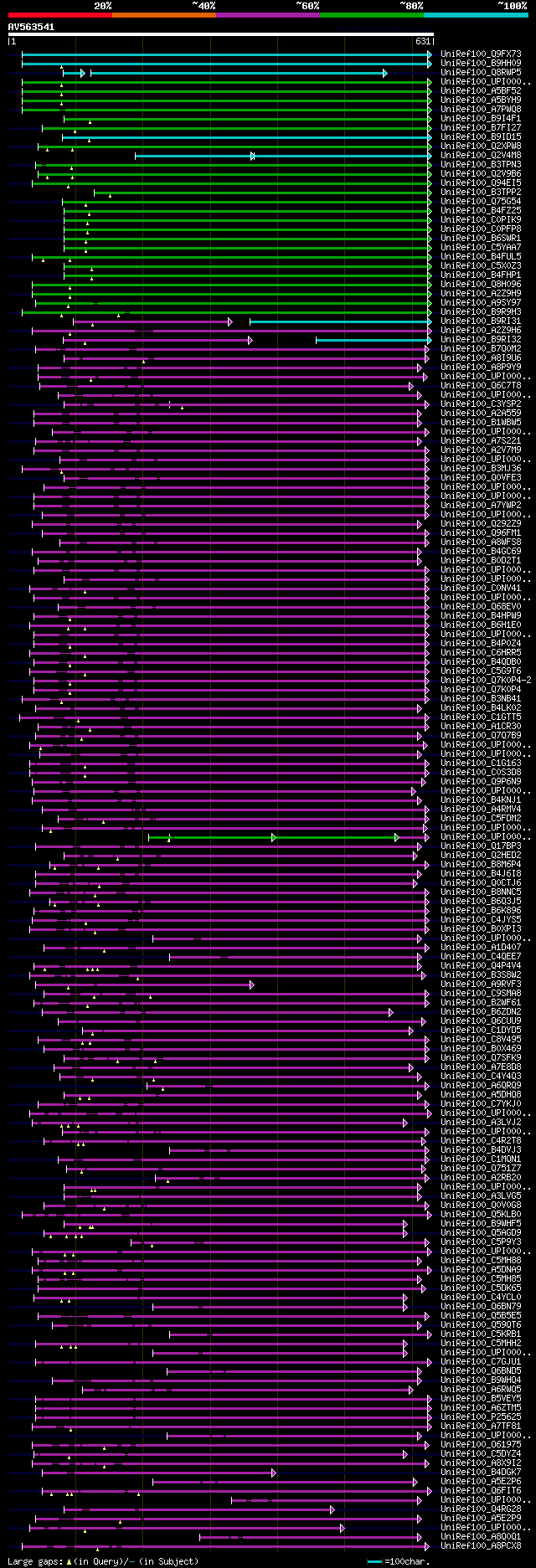
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV563541 SQ189b06F
(631 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q9FX73 AT1G16560 protein n=2 Tax=Arabidopsis thaliana ... 441 e-122
UniRef100_B9HH09 Predicted protein n=1 Tax=Populus trichocarpa R... 340 6e-92
UniRef100_Q8RWP5 Putative uncharacterized protein At1g16560 n=1 ... 327 4e-88
UniRef100_UPI00019856E6 PREDICTED: hypothetical protein isoform ... 322 2e-86
UniRef100_A5BF52 Putative uncharacterized protein n=1 Tax=Vitis ... 322 2e-86
UniRef100_A5BYH9 Chromosome chr16 scaffold_189, whole genome sho... 308 3e-82
UniRef100_A7PWQ8 Chromosome chr19 scaffold_35, whole genome shot... 304 5e-81
UniRef100_B9I4F1 Predicted protein n=2 Tax=Populus RepID=B9I4F1_... 292 2e-77
UniRef100_B7FI27 Putative uncharacterized protein n=1 Tax=Medica... 291 2e-77
UniRef100_B9ID15 Predicted protein n=1 Tax=Populus trichocarpa R... 291 3e-77
UniRef100_Q2XPW8 Per1-like family protein n=1 Tax=Solanum tubero... 286 1e-75
UniRef100_Q2V4M8 Putative uncharacterized protein At1g16560.4 n=... 186 3e-75
UniRef100_B3TPN3 PERLD1 n=1 Tax=Glycine max RepID=B3TPN3_SOYBN 283 8e-75
UniRef100_Q2V9B6 Per1-like family protein n=1 Tax=Solanum tubero... 283 1e-74
UniRef100_Q94EI5 AT5g62130/mtg10_150 n=2 Tax=Arabidopsis thalian... 271 4e-71
UniRef100_B3TPP2 PERLD1 (Fragment) n=1 Tax=Solanum commersonii R... 261 3e-68
UniRef100_Q75G54 Os05g0220100 protein n=2 Tax=Oryza sativa RepID... 255 2e-66
UniRef100_B4FZ25 Putative uncharacterized protein n=1 Tax=Zea ma... 250 6e-65
UniRef100_C0PIK9 Putative uncharacterized protein n=1 Tax=Zea ma... 249 1e-64
UniRef100_C0PFP8 Putative uncharacterized protein n=1 Tax=Zea ma... 249 1e-64
UniRef100_B6SWR1 CAB2 n=1 Tax=Zea mays RepID=B6SWR1_MAIZE 249 1e-64
UniRef100_C5YAA7 Putative uncharacterized protein Sb06g019590 n=... 248 2e-64
UniRef100_B4FUL5 Putative uncharacterized protein n=1 Tax=Zea ma... 237 5e-61
UniRef100_C5X0Z3 Putative uncharacterized protein Sb01g049340 n=... 236 9e-61
UniRef100_B4FHP1 Putative uncharacterized protein n=2 Tax=Zea ma... 232 2e-59
UniRef100_Q8H096 Os10g0524100 protein n=1 Tax=Oryza sativa Japon... 231 3e-59
UniRef100_A2Z9H9 Putative uncharacterized protein n=1 Tax=Oryza ... 230 6e-59
UniRef100_A9SY97 Predicted protein n=1 Tax=Physcomitrella patens... 226 1e-57
UniRef100_B9R9H3 Putative uncharacterized protein n=1 Tax=Ricinu... 223 1e-56
UniRef100_B9RI31 Putative uncharacterized protein n=1 Tax=Ricinu... 166 1e-45
UniRef100_A2Z9H6 Putative uncharacterized protein n=1 Tax=Oryza ... 172 3e-41
UniRef100_B9RI32 Putative uncharacterized protein n=1 Tax=Ricinu... 103 4e-32
UniRef100_B7Q0M2 Post-GPI attachment to proteins factor, putativ... 128 5e-28
UniRef100_A8I9U6 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 127 8e-28
UniRef100_A8P9Y9 Putative uncharacterized protein n=1 Tax=Coprin... 126 1e-27
UniRef100_UPI0001792D40 PREDICTED: similar to CG3271 CG3271-PA n... 124 5e-27
UniRef100_Q6C7T8 YALI0D25454p n=1 Tax=Yarrowia lipolytica RepID=... 122 2e-26
UniRef100_UPI0001758763 PREDICTED: similar to CG3271 CG3271-PB n... 122 3e-26
UniRef100_C3YSP2 Putative uncharacterized protein (Fragment) n=1... 120 9e-26
UniRef100_A2A559 Post-GPI attachment to proteins factor 3 n=1 Ta... 119 3e-25
UniRef100_B1WBW5 Perld1 protein n=1 Tax=Rattus norvegicus RepID=... 118 4e-25
UniRef100_UPI000175FE92 PREDICTED: similar to CAB2 protein n=1 T... 118 5e-25
UniRef100_A7S221 Predicted protein n=1 Tax=Nematostella vectensi... 118 5e-25
UniRef100_A2V7M9 Post-GPI attachment to proteins factor 3 n=1 Ta... 117 1e-24
UniRef100_UPI0001A2C528 UPI0001A2C528 related cluster n=1 Tax=Da... 116 1e-24
UniRef100_B3MJ36 GF11063 n=1 Tax=Drosophila ananassae RepID=B3MJ... 115 2e-24
UniRef100_Q0VFE3 Post-GPI attachment to proteins factor 3 n=1 Ta... 115 2e-24
UniRef100_UPI00016E9E6F UPI00016E9E6F related cluster n=1 Tax=Ta... 115 3e-24
UniRef100_UPI00006147F7 UPI00006147F7 related cluster n=1 Tax=Bo... 115 3e-24
UniRef100_A7YWP2 Post-GPI attachment to proteins factor 3 n=1 Ta... 115 3e-24
UniRef100_UPI0000D9E34D PREDICTED: similar to CAB2 protein isofo... 115 4e-24
UniRef100_Q292Z9 GA17095 n=1 Tax=Drosophila pseudoobscura pseudo... 115 4e-24
UniRef100_Q96FM1 Post-GPI attachment to proteins factor 3 n=1 Ta... 115 4e-24
UniRef100_A8WFS8 Post-GPI attachment to proteins factor 3 n=1 Ta... 115 4e-24
UniRef100_B4GC69 GL11111 n=1 Tax=Drosophila persimilis RepID=B4G... 114 5e-24
UniRef100_B0D2T1 Predicted protein n=1 Tax=Laccaria bicolor S238... 114 5e-24
UniRef100_UPI00005A1C0B PREDICTED: similar to CAB2 protein n=1 T... 114 7e-24
UniRef100_UPI00017B3C98 UPI00017B3C98 related cluster n=1 Tax=Te... 114 7e-24
UniRef100_C0NV41 PER1 n=1 Tax=Ajellomyces capsulatus G186AR RepI... 114 9e-24
UniRef100_UPI000155F217 PREDICTED: similar to CAB2 protein n=1 T... 113 1e-23
UniRef100_Q68EV0 Post-GPI attachment to proteins factor 3 n=1 Ta... 113 1e-23
UniRef100_B4HPW9 GM20888 n=1 Tax=Drosophila sechellia RepID=B4HP... 112 2e-23
UniRef100_B6H1E0 Pc13g02730 protein n=1 Tax=Penicillium chrysoge... 112 3e-23
UniRef100_UPI00005E9C97 PREDICTED: similar to Per1-like domain c... 112 3e-23
UniRef100_B4P0Z4 GE19068 n=1 Tax=Drosophila yakuba RepID=B4P0Z4_... 112 3e-23
UniRef100_C6HRR5 Mn2+ homeostasis protein n=1 Tax=Ajellomyces ca... 111 4e-23
UniRef100_B4QDB0 GD10418 n=1 Tax=Drosophila simulans RepID=B4QDB... 111 6e-23
UniRef100_C5G9T6 Mn2+ homeostasis protein n=2 Tax=Ajellomyces de... 111 6e-23
UniRef100_Q7K0P4-2 Isoform 2 of Post-GPI attachment to proteins ... 111 6e-23
UniRef100_Q7K0P4 Post-GPI attachment to proteins factor 3 n=1 Ta... 111 6e-23
UniRef100_B3NB41 GG23214 n=1 Tax=Drosophila erecta RepID=B3NB41_... 110 7e-23
UniRef100_B4LK02 GJ22170 n=1 Tax=Drosophila virilis RepID=B4LK02... 110 1e-22
UniRef100_C1GTT5 Mn2+ homeostasis protein (Per1) n=1 Tax=Paracoc... 110 1e-22
UniRef100_A1CR30 Mn2+ homeostasis protein (Per1), putative n=1 T... 110 1e-22
UniRef100_Q7Q7B9 AGAP005392-PA n=1 Tax=Anopheles gambiae RepID=Q... 109 2e-22
UniRef100_UPI000180CEE0 PREDICTED: similar to CAB2 protein n=1 T... 109 2e-22
UniRef100_UPI0000DB769A PREDICTED: similar to CG3271-PB, isoform... 109 2e-22
UniRef100_C1G163 Mn2+ homeostasis protein (Per1) n=1 Tax=Paracoc... 109 2e-22
UniRef100_C0S3D8 Putative uncharacterized protein n=1 Tax=Paraco... 109 2e-22
UniRef100_Q9P6N9 Protein PER1 homolog n=1 Tax=Schizosaccharomyce... 109 2e-22
UniRef100_UPI0000EB20C6 CAB2 protein n=1 Tax=Canis lupus familia... 108 4e-22
UniRef100_B4KNJ1 GI19294 n=1 Tax=Drosophila mojavensis RepID=B4K... 107 6e-22
UniRef100_A4RMV4 Putative uncharacterized protein n=1 Tax=Magnap... 107 6e-22
UniRef100_C5FDM2 Putative uncharacterized protein n=1 Tax=Micros... 107 8e-22
UniRef100_UPI000186E4AA conserved hypothetical protein n=1 Tax=P... 106 1e-21
UniRef100_UPI0001860101 hypothetical protein BRAFLDRAFT_67792 n=... 106 1e-21
UniRef100_Q17BP3 Putative uncharacterized protein n=1 Tax=Aedes ... 106 1e-21
UniRef100_Q2HED2 Putative uncharacterized protein n=1 Tax=Chaeto... 106 1e-21
UniRef100_B8M6P4 Mn2 homeostasis protein (Per1), putative n=1 Ta... 106 1e-21
UniRef100_B4J6I8 GH21747 n=1 Tax=Drosophila grimshawi RepID=B4J6... 106 2e-21
UniRef100_Q0CTJ6 Putative uncharacterized protein n=1 Tax=Asperg... 106 2e-21
UniRef100_B8NNC5 Mn2+ homeostasis protein (Per1), putative n=1 T... 106 2e-21
UniRef100_B6Q3J5 Mn2+ homeostasis protein (Per1), putative n=1 T... 106 2e-21
UniRef100_B6K896 PER1 n=1 Tax=Schizosaccharomyces japonicus yFS2... 106 2e-21
UniRef100_C4JYS5 Putative uncharacterized protein n=1 Tax=Uncino... 105 3e-21
UniRef100_B0XPI3 Mn2+ homeostasis protein (Per1), putative n=2 T... 105 3e-21
UniRef100_UPI00015B4CA5 PREDICTED: similar to LD44494p n=1 Tax=N... 104 5e-21
UniRef100_A1D407 Mn2+ homeostasis protein (Per1), putative n=1 T... 104 5e-21
UniRef100_C4QEE7 Expressed protein n=1 Tax=Schistosoma mansoni R... 102 3e-20
UniRef100_Q4P4V4 Putative uncharacterized protein n=1 Tax=Ustila... 102 3e-20
UniRef100_B3S8W2 Putative uncharacterized protein n=1 Tax=Tricho... 102 3e-20
UniRef100_A9RVF3 Predicted protein n=1 Tax=Physcomitrella patens... 101 5e-20
UniRef100_C9SMA8 PER1 n=1 Tax=Verticillium albo-atrum VaMs.102 R... 101 5e-20
UniRef100_B2WF61 Mn2+ homeostasis protein Per1 n=1 Tax=Pyrenopho... 101 5e-20
UniRef100_B6ZDN2 Putative uncharacterized protein PGAP3 n=1 Tax=... 101 6e-20
UniRef100_Q6CUU9 KLLA0C02101p n=1 Tax=Kluyveromyces lactis RepID... 101 6e-20
UniRef100_C1DYD5 Per1-like family protein n=1 Tax=Micromonas sp.... 100 8e-20
UniRef100_C8V495 Mn2+ homeostasis protein (Per1), putative (AFU_... 100 1e-19
UniRef100_B0X469 Putative uncharacterized protein n=1 Tax=Culex ... 100 1e-19
UniRef100_Q7SFK9 Putative uncharacterized protein n=1 Tax=Neuros... 99 3e-19
UniRef100_A7E8D8 Putative uncharacterized protein n=1 Tax=Sclero... 99 3e-19
UniRef100_C4Y4Q3 Putative uncharacterized protein n=1 Tax=Clavis... 98 5e-19
UniRef100_A6QRQ9 Putative uncharacterized protein n=1 Tax=Ajello... 96 3e-18
UniRef100_A5DHQ8 Putative uncharacterized protein n=1 Tax=Pichia... 95 6e-18
UniRef100_C7YKJ0 Predicted protein n=1 Tax=Nectria haematococca ... 94 7e-18
UniRef100_UPI0000589060 PREDICTED: similar to MGC84367 protein n... 94 1e-17
UniRef100_A3LVJ2 Predicted protein n=1 Tax=Pichia stipitis RepID... 94 1e-17
UniRef100_UPI000023E7E4 hypothetical protein FG01138.1 n=1 Tax=G... 93 2e-17
UniRef100_C4R2T8 Protein of the endoplasmic reticulum, required ... 93 2e-17
UniRef100_B4DVJ3 cDNA FLJ61173, highly similar to Homo sapiens p... 92 4e-17
UniRef100_C1MQN1 Per1-like family protein n=1 Tax=Micromonas pus... 92 5e-17
UniRef100_Q751Z7 AFR678Cp n=1 Tax=Eremothecium gossypii RepID=Q7... 92 5e-17
UniRef100_A2RB20 Similarity to hypothetical membrane protein YCR... 91 1e-16
UniRef100_UPI000151B203 hypothetical protein PGUG_02809 n=1 Tax=... 90 1e-16
UniRef100_A3LVG5 Predicted protein n=1 Tax=Pichia stipitis RepID... 90 2e-16
UniRef100_Q0V0G8 Putative uncharacterized protein n=1 Tax=Phaeos... 89 4e-16
UniRef100_Q5KLB0 Manganese ion homeostasis-related protein, puta... 88 5e-16
UniRef100_B9WHF5 ER protein processing protein, putative n=1 Tax... 88 5e-16
UniRef100_Q5AGD9 Putative uncharacterized protein n=1 Tax=Candid... 88 7e-16
UniRef100_C5P9Y3 Per1-like family protein n=1 Tax=Coccidioides p... 88 7e-16
UniRef100_UPI000151B382 hypothetical protein PGUG_04760 n=1 Tax=... 87 9e-16
UniRef100_C5MH88 Putative uncharacterized protein n=1 Tax=Candid... 87 9e-16
UniRef100_A5DNA9 Putative uncharacterized protein n=1 Tax=Pichia... 87 9e-16
UniRef100_C5MH85 Putative uncharacterized protein n=1 Tax=Candid... 86 2e-15
UniRef100_C5DK65 KLTH0F02134p n=1 Tax=Lachancea thermotolerans C... 86 3e-15
UniRef100_C4YCL0 Putative uncharacterized protein n=1 Tax=Clavis... 86 3e-15
UniRef100_Q6BN79 DEHA2E23936p n=1 Tax=Debaryomyces hansenii RepI... 86 3e-15
UniRef100_Q5B5E5 Putative uncharacterized protein n=1 Tax=Emeric... 85 6e-15
UniRef100_Q59QT6 Putative uncharacterized protein n=1 Tax=Candid... 85 6e-15
UniRef100_C5KRB1 Putative uncharacterized protein n=1 Tax=Perkin... 84 7e-15
UniRef100_C5MHH2 Putative uncharacterized protein n=1 Tax=Candid... 84 7e-15
UniRef100_UPI00003BDFC8 hypothetical protein DEHA0E25366g n=1 Ta... 84 1e-14
UniRef100_C7GJU1 Per1p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 84 1e-14
UniRef100_Q6BND5 DEHA2E22638p n=1 Tax=Debaryomyces hansenii RepI... 83 2e-14
UniRef100_B9WHQ4 ER protein processing protein, putative (Er pro... 83 2e-14
UniRef100_A6RWQ5 Putative uncharacterized protein n=1 Tax=Botryo... 82 4e-14
UniRef100_B5VEY5 YCR044Cp-like protein (Fragment) n=1 Tax=Saccha... 82 5e-14
UniRef100_A6ZTM5 Vacuolar membrane protein n=3 Tax=Saccharomyces... 82 5e-14
UniRef100_P25625 Protein PER1 n=1 Tax=Saccharomyces cerevisiae R... 82 5e-14
UniRef100_A7TF81 Putative uncharacterized protein n=1 Tax=Vander... 81 8e-14
UniRef100_UPI00003BDF90 hypothetical protein DEHA0E24024g n=1 Ta... 79 2e-13
UniRef100_O61975 Putative uncharacterized protein R01B10.4 n=1 T... 79 4e-13
UniRef100_C5DYZ4 ZYRO0F16940p n=2 Tax=Zygosaccharomyces rouxii R... 78 7e-13
UniRef100_A8X9I2 Putative uncharacterized protein n=1 Tax=Caenor... 77 1e-12
UniRef100_B4DGK7 cDNA FLJ52489, highly similar to Homo sapiens p... 75 3e-12
UniRef100_A5E2P6 Putative uncharacterized protein n=1 Tax=Lodder... 74 1e-11
UniRef100_Q6FIT6 Strain CBS138 chromosome M complete sequence n=... 73 2e-11
UniRef100_UPI0000ECBFBE CAB2 protein n=1 Tax=Gallus gallus RepID... 72 3e-11
UniRef100_Q4RG28 Chromosome 2 SCAF15106, whole genome shotgun se... 72 3e-11
UniRef100_A5E2P9 Putative uncharacterized protein n=1 Tax=Lodder... 70 1e-10
UniRef100_UPI0000588E78 PREDICTED: hypothetical protein, partial... 67 2e-09
UniRef100_A8Q0Q1 Putative uncharacterized protein n=1 Tax=Malass... 61 7e-08
UniRef100_A8PCX8 Per1-like family protein n=1 Tax=Brugia malayi ... 59 4e-07
UniRef100_Q8K2Q4 Perld1 protein (Fragment) n=1 Tax=Mus musculus ... 56 3e-06