[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
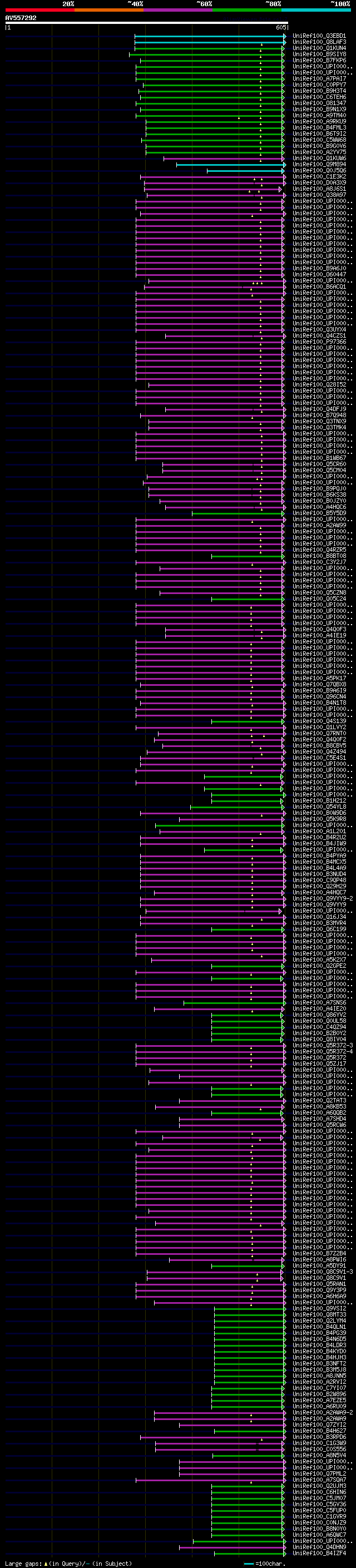
Query= AV557292 SQ064h04F
(605 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q3EBD1 Putative uncharacterized protein At3g02460.2 n=... 221 3e-56
UniRef100_Q8LAF3 Putative plant adhesion molecule n=1 Tax=Arabid... 209 1e-52
UniRef100_Q1KUN4 Putative uncharacterized protein n=1 Tax=Cleome... 180 9e-44
UniRef100_B9SIY8 Ecotropic viral integration site, putative n=1 ... 175 2e-42
UniRef100_B7FKP6 Putative uncharacterized protein n=1 Tax=Medica... 173 1e-41
UniRef100_UPI0001984B69 PREDICTED: hypothetical protein isoform ... 172 2e-41
UniRef100_UPI0001984B68 PREDICTED: hypothetical protein isoform ... 172 2e-41
UniRef100_A7PAI7 Chromosome chr14 scaffold_9, whole genome shotg... 172 2e-41
UniRef100_C0PPY7 Putative uncharacterized protein n=1 Tax=Picea ... 169 2e-40
UniRef100_B9H3T4 Predicted protein (Fragment) n=2 Tax=Populus tr... 167 5e-40
UniRef100_C6TEH6 Putative uncharacterized protein n=1 Tax=Glycin... 166 2e-39
UniRef100_O81347 Plant adhesion molecule 1 n=2 Tax=Arabidopsis t... 163 9e-39
UniRef100_B9N1X9 Predicted protein n=1 Tax=Populus trichocarpa R... 159 2e-37
UniRef100_A9TM40 Predicted protein n=1 Tax=Physcomitrella patens... 157 8e-37
UniRef100_A9RKU9 Predicted protein n=1 Tax=Physcomitrella patens... 155 3e-36
UniRef100_B4FML3 Putative uncharacterized protein n=2 Tax=Zea ma... 150 8e-35
UniRef100_B6T9I2 USP6 N-terminal-like protein n=1 Tax=Zea mays R... 149 2e-34
UniRef100_C5WW68 Putative uncharacterized protein Sb01g004460 n=... 149 2e-34
UniRef100_B9G0V6 Putative uncharacterized protein n=1 Tax=Oryza ... 144 4e-33
UniRef100_A2YV75 Putative uncharacterized protein n=1 Tax=Oryza ... 144 5e-33
UniRef100_Q1KUW6 Putative uncharacterized protein n=1 Tax=Cleome... 136 1e-30
UniRef100_Q9M894 Putative plant adhesion molecule n=1 Tax=Arabid... 117 9e-25
UniRef100_Q0J5Q6 Os08g0412600 protein n=1 Tax=Oryza sativa Japon... 94 8e-18
UniRef100_C1E3K2 Predicted protein n=1 Tax=Micromonas sp. RCC299... 84 9e-15
UniRef100_D0A3X9 GTPase activating protein, putative n=1 Tax=Try... 77 1e-12
UniRef100_A8J6S1 RabGAP/TBC protein n=1 Tax=Chlamydomonas reinha... 76 2e-12
UniRef100_Q38A97 GTPase activating protein, putative n=1 Tax=Try... 75 3e-12
UniRef100_UPI00017EF9C1 PREDICTED: similar to Ecotropic viral in... 71 8e-11
UniRef100_UPI00017C2BED PREDICTED: similar to Ecotropic viral in... 71 8e-11
UniRef100_UPI0001757F72 PREDICTED: similar to GA11164-PA n=1 Tax... 71 8e-11
UniRef100_UPI0000E1EA5B PREDICTED: ecotropic viral integration s... 71 8e-11
UniRef100_UPI0000E1EA5A PREDICTED: ecotropic viral integration s... 71 8e-11
UniRef100_UPI0000E1EA59 PREDICTED: hypothetical protein isoform ... 71 8e-11
UniRef100_UPI0000D99A2E PREDICTED: similar to ecotropic viral in... 71 8e-11
UniRef100_UPI00005A128C PREDICTED: similar to ecotropic viral in... 71 8e-11
UniRef100_UPI0000EB3692 Ecotropic viral integration site 5 prote... 71 8e-11
UniRef100_UPI0000EB3691 Ecotropic viral integration site 5 prote... 71 8e-11
UniRef100_UPI0000EB368F Ecotropic viral integration site 5 prote... 71 8e-11
UniRef100_B9A6J0 Ecotropic viral integration site 5 n=1 Tax=Homo... 70 1e-10
UniRef100_O60447 Ecotropic viral integration site 5 protein homo... 70 1e-10
UniRef100_UPI00015B61A7 PREDICTED: similar to ENSANGP00000001286... 70 1e-10
UniRef100_B6ACQ1 TBC domain-containing protein n=1 Tax=Cryptospo... 70 1e-10
UniRef100_UPI0001865795 hypothetical protein BRAFLDRAFT_88764 n=... 70 2e-10
UniRef100_UPI0001B7A5FC UPI0001B7A5FC related cluster n=1 Tax=Ra... 70 2e-10
UniRef100_UPI0000DC12D4 ecotropic viral integration site 5 n=1 T... 70 2e-10
UniRef100_UPI000021D659 ecotropic viral integration site 5 n=1 T... 70 2e-10
UniRef100_UPI00005642BC ecotropic viral integration site 5 n=1 T... 70 2e-10
UniRef100_UPI00001929A5 ecotropic viral integration site 5 n=1 T... 70 2e-10
UniRef100_Q3UYX4 Putative uncharacterized protein (Fragment) n=1... 70 2e-10
UniRef100_Q4CZS1 Rab-like GTPase activating protein, putative n=... 70 2e-10
UniRef100_P97366 Ecotropic viral integration site 5 protein n=1 ... 70 2e-10
UniRef100_UPI000194CBFB PREDICTED: ecotropic viral integration s... 69 2e-10
UniRef100_UPI0000E80A62 PREDICTED: hypothetical protein n=1 Tax=... 69 2e-10
UniRef100_UPI00016E912C UPI00016E912C related cluster n=1 Tax=Ta... 69 2e-10
UniRef100_UPI00016E912B UPI00016E912B related cluster n=1 Tax=Ta... 69 2e-10
UniRef100_UPI00016E912A UPI00016E912A related cluster n=1 Tax=Ta... 69 2e-10
UniRef100_UPI0000ECB2FA Ecotropic viral integration site 5 prote... 69 2e-10
UniRef100_Q28I52 Novel protein simiar to eiv5 (Fragment) n=1 Tax... 69 2e-10
UniRef100_UPI000175F8A6 PREDICTED: similar to Ecotropic viral in... 69 3e-10
UniRef100_UPI000155D268 PREDICTED: hypothetical protein, partial... 69 3e-10
UniRef100_UPI000054609D PREDICTED: ecotropic viral integration s... 69 3e-10
UniRef100_Q4DFJ9 Rab-like GTPase activating protein, putative n=... 69 3e-10
UniRef100_B7Q948 Ecotropic viral integration site, putative n=1 ... 69 4e-10
UniRef100_Q3TNX9 Putative uncharacterized protein (Fragment) n=1... 68 5e-10
UniRef100_Q3TMK4 Putative uncharacterized protein (Fragment) n=1... 68 5e-10
UniRef100_UPI000069F32D EVI5-like protein (Ecotropic viral integ... 67 1e-09
UniRef100_UPI000069F32C EVI5-like protein (Ecotropic viral integ... 67 1e-09
UniRef100_UPI000069F32B EVI5-like protein (Ecotropic viral integ... 67 1e-09
UniRef100_UPI000069F32A EVI5-like protein (Ecotropic viral integ... 67 1e-09
UniRef100_B1WB67 LOC100145757 protein n=1 Tax=Xenopus (Silurana)... 67 1e-09
UniRef100_Q5CR60 TBC domain containing protein n=1 Tax=Cryptospo... 66 2e-09
UniRef100_Q5CM04 Putative uncharacterized protein n=1 Tax=Crypto... 66 2e-09
UniRef100_UPI000186ECD1 GTPase-activating protein GYP5, putative... 66 2e-09
UniRef100_UPI00017B4253 UPI00017B4253 related cluster n=1 Tax=Te... 66 2e-09
UniRef100_B9PQJ0 TBC domain-containing protein, putative n=2 Tax... 66 2e-09
UniRef100_B6KS38 TBC domain-containing protein n=1 Tax=Toxoplasm... 66 2e-09
UniRef100_B0JZY0 LOC733501 protein (Fragment) n=1 Tax=Xenopus (S... 65 4e-09
UniRef100_A4HQC6 Rab-like GTPase activating protein, putative n=... 64 7e-09
UniRef100_B5Y5D9 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 64 9e-09
UniRef100_UPI00018699E7 hypothetical protein BRAFLDRAFT_251188 n... 64 1e-08
UniRef100_A2AW99 Novel protein similar to human and mouse ecotro... 64 1e-08
UniRef100_UPI00017B203D UPI00017B203D related cluster n=1 Tax=Te... 64 1e-08
UniRef100_UPI00017B203C UPI00017B203C related cluster n=1 Tax=Te... 64 1e-08
UniRef100_UPI00017B203B UPI00017B203B related cluster n=1 Tax=Te... 64 1e-08
UniRef100_Q4RZR5 Chromosome 18 SCAF14786, whole genome shotgun s... 64 1e-08
UniRef100_B8BT08 Predicted protein n=1 Tax=Thalassiosira pseudon... 64 1e-08
UniRef100_C3Y2J7 Putative uncharacterized protein n=1 Tax=Branch... 64 1e-08
UniRef100_UPI0000569D12 LOC553306 protein n=1 Tax=Danio rerio Re... 63 2e-08
UniRef100_UPI00016E2DE0 UPI00016E2DE0 related cluster n=1 Tax=Ta... 63 2e-08
UniRef100_UPI00016E2DDE UPI00016E2DDE related cluster n=1 Tax=Ta... 63 2e-08
UniRef100_UPI00016E2DDD UPI00016E2DDD related cluster n=1 Tax=Ta... 63 2e-08
UniRef100_Q5CZN8 Evi5 protein (Fragment) n=1 Tax=Danio rerio Rep... 63 2e-08
UniRef100_Q05C24 Evi5 protein (Fragment) n=1 Tax=Mus musculus Re... 63 2e-08
UniRef100_UPI0001796425 PREDICTED: ecotropic viral integration s... 63 2e-08
UniRef100_UPI0000EB1983 EVI5-like protein (Ecotropic viral integ... 63 2e-08
UniRef100_UPI0000EB1982 EVI5-like protein (Ecotropic viral integ... 63 2e-08
UniRef100_UPI00004BD92B PREDICTED: similar to CG11727-PA, isofor... 63 2e-08
UniRef100_Q4Q0F3 Rab-like GTPase activating protein, putative n=... 63 2e-08
UniRef100_A4IE19 Rab-like GTPase activating protein, putative n=... 63 2e-08
UniRef100_UPI0000D9EA0C PREDICTED: similar to ecotropic viral in... 62 3e-08
UniRef100_UPI0001B7990D RIKEN cDNA B130050I23 gene n=1 Tax=Rattu... 62 3e-08
UniRef100_UPI00015DEEA2 RIKEN cDNA B130050I23 gene n=1 Tax=Mus m... 62 3e-08
UniRef100_UPI0000DBD689 ecotropic viral integration site 5 like ... 62 3e-08
UniRef100_UPI000179EA3A UPI000179EA3A related cluster n=1 Tax=Bo... 62 3e-08
UniRef100_UPI000179EA39 UPI000179EA39 related cluster n=1 Tax=Bo... 62 3e-08
UniRef100_A5PK17 EVI5L protein n=1 Tax=Bos taurus RepID=A5PK17_B... 62 3e-08
UniRef100_Q7QBX8 AGAP002354-PA (Fragment) n=1 Tax=Anopheles gamb... 62 3e-08
UniRef100_B9A6I9 Ecotropic viral integration site 5-like n=1 Tax... 62 3e-08
UniRef100_Q96CN4 EVI5-like protein n=1 Tax=Homo sapiens RepID=EV... 62 3e-08
UniRef100_B4N1T8 GK16367 n=1 Tax=Drosophila willistoni RepID=B4N... 62 4e-08
UniRef100_UPI0000F2120F PREDICTED: novel GTPase activating prote... 62 5e-08
UniRef100_UPI0001A2BE61 Novel GTPase activating protein n=1 Tax=... 62 5e-08
UniRef100_Q4S139 Chromosome 1 SCAF14770, whole genome shotgun se... 62 5e-08
UniRef100_Q1LVY2 Novel GTPase activating protein (Fragment) n=1 ... 62 5e-08
UniRef100_Q7RNT0 Plant adhesion molecule 1-related n=1 Tax=Plasm... 62 5e-08
UniRef100_Q4Q0F2 Rab-like GTPase activating protein, putative n=... 62 5e-08
UniRef100_B8CBV5 RabGAP (Fragment) n=1 Tax=Thalassiosira pseudon... 61 6e-08
UniRef100_Q4Z494 Putative uncharacterized protein (Fragment) n=1... 61 6e-08
UniRef100_C5E4S1 ZYRO0E08316p n=1 Tax=Zygosaccharomyces rouxii C... 61 6e-08
UniRef100_UPI0001793849 PREDICTED: similar to AGAP002354-PA, par... 61 8e-08
UniRef100_UPI0000F2CABB PREDICTED: hypothetical protein n=1 Tax=... 61 8e-08
UniRef100_UPI00005A36B8 PREDICTED: similar to TBC1 domain family... 61 8e-08
UniRef100_UPI0001A2D1F1 UPI0001A2D1F1 related cluster n=1 Tax=Da... 61 8e-08
UniRef100_UPI00004BCE1B PREDICTED: similar to TBC1 domain family... 61 8e-08
UniRef100_UPI000069F329 EVI5-like protein (Ecotropic viral integ... 60 1e-07
UniRef100_B1H212 TBC1 domain family, member 10C n=1 Tax=Rattus n... 60 1e-07
UniRef100_Q54YL8 Ankyrin repeat-containing protein n=1 Tax=Dicty... 60 1e-07
UniRef100_B0W9D6 Ecotropic viral integration site n=1 Tax=Culex ... 60 1e-07
UniRef100_Q5K9R8 Putative uncharacterized protein n=1 Tax=Filoba... 60 1e-07
UniRef100_UPI0000E4947B PREDICTED: similar to ecotropic viral in... 60 1e-07
UniRef100_A1L201 Evi5 protein (Fragment) n=1 Tax=Danio rerio Rep... 60 1e-07
UniRef100_B4R2U2 GD15988 n=1 Tax=Drosophila simulans RepID=B4R2U... 60 1e-07
UniRef100_B4JIW9 GH12377 n=1 Tax=Drosophila grimshawi RepID=B4JI... 60 1e-07
UniRef100_UPI0001796DEA PREDICTED: TBC1 domain family, member 10... 60 2e-07
UniRef100_B4PYA9 GE17295 n=1 Tax=Drosophila yakuba RepID=B4PYA9_... 60 2e-07
UniRef100_B4MCX5 GJ15325 n=1 Tax=Drosophila virilis RepID=B4MCX5... 60 2e-07
UniRef100_B4L4A9 GI14892 n=1 Tax=Drosophila mojavensis RepID=B4L... 60 2e-07
UniRef100_B3NUD4 GG18855 n=1 Tax=Drosophila erecta RepID=B3NUD4_... 60 2e-07
UniRef100_C9QP48 MIP13364p n=1 Tax=Drosophila melanogaster RepID... 59 2e-07
UniRef100_Q29H29 GA11164 n=2 Tax=pseudoobscura subgroup RepID=Q2... 59 2e-07
UniRef100_A4HQC7 Rab-like GTPase activating protein, putative n=... 59 2e-07
UniRef100_Q9VYY9-2 Isoform B of TBC1 domain family member CG1172... 59 2e-07
UniRef100_Q9VYY9 TBC1 domain family member CG11727 n=1 Tax=Droso... 59 2e-07
UniRef100_UPI0000E4616F PREDICTED: similar to mKIAA1055 protein ... 59 3e-07
UniRef100_Q16J34 Ecotropic viral integration site n=1 Tax=Aedes ... 59 3e-07
UniRef100_B3MVR4 GF22394 n=1 Tax=Drosophila ananassae RepID=B3MV... 59 3e-07
UniRef100_Q6C199 YALI0F18106p n=1 Tax=Yarrowia lipolytica RepID=... 59 3e-07
UniRef100_UPI000194CABA PREDICTED: RAB GTPase activating protein... 59 4e-07
UniRef100_UPI00005A1F66 PREDICTED: similar to RAB GTPase activat... 59 4e-07
UniRef100_UPI0000EB2355 RAB GTPase activating protein 1 n=3 Tax=... 59 4e-07
UniRef100_UPI0000ECBE92 EVI5-like protein (Ecotropic viral integ... 59 4e-07
UniRef100_A5K2X7 TBC domain containing protein n=1 Tax=Plasmodiu... 59 4e-07
UniRef100_Q2GPE2 Putative uncharacterized protein n=1 Tax=Chaeto... 59 4e-07
UniRef100_UPI0000E1EE1F PREDICTED: hypothetical protein n=1 Tax=... 58 5e-07
UniRef100_UPI0000D9D724 PREDICTED: similar to TBC1 domain family... 58 5e-07
UniRef100_UPI0000ECB287 RAB GTPase activating protein 1-like n=1... 58 5e-07
UniRef100_UPI0000ECB286 RAB GTPase activating protein 1-like n=1... 58 5e-07
UniRef100_UPI0000ECB285 RAB GTPase activating protein 1-like n=1... 58 5e-07
UniRef100_A7SNS6 Predicted protein n=1 Tax=Nematostella vectensi... 58 5e-07
UniRef100_A4IE20 Rab-like GTPase activating protein, putative n=... 58 5e-07
UniRef100_Q86YV2 FLJ00332 protein (Fragment) n=1 Tax=Homo sapien... 58 5e-07
UniRef100_Q0UL58 Putative uncharacterized protein n=1 Tax=Phaeos... 58 5e-07
UniRef100_C4QZ94 GTPase-activating protein n=1 Tax=Pichia pastor... 58 5e-07
UniRef100_B2B0Y2 Predicted CDS Pa_3_7710 (Fragment) n=1 Tax=Podo... 58 5e-07
UniRef100_Q8IV04 Carabin n=1 Tax=Homo sapiens RepID=TB10C_HUMAN 58 5e-07
UniRef100_Q5R372-3 Isoform 3 of RAB GTPase-activating protein 1-... 58 5e-07
UniRef100_Q5R372-4 Isoform 4 of RAB GTPase-activating protein 1-... 58 5e-07
UniRef100_Q5R372 RAB GTPase-activating protein 1-like n=2 Tax=Ho... 58 5e-07
UniRef100_Q5ZJ17 RAB GTPase-activating protein 1-like n=1 Tax=Ga... 58 5e-07
UniRef100_UPI000192481E PREDICTED: similar to predicted protein ... 58 7e-07
UniRef100_UPI000069D92D UPI000069D92D related cluster n=1 Tax=Xe... 58 7e-07
UniRef100_UPI00016E240F UPI00016E240F related cluster n=1 Tax=Ta... 58 7e-07
UniRef100_UPI000179D0EB UPI000179D0EB related cluster n=1 Tax=Bo... 58 7e-07
UniRef100_UPI000179D0EA UPI000179D0EA related cluster n=1 Tax=Bo... 58 7e-07
UniRef100_Q2TAT3 MGC130926 protein n=1 Tax=Xenopus laevis RepID=... 58 7e-07
UniRef100_A8KB53 LOC566318 protein (Fragment) n=1 Tax=Danio reri... 58 7e-07
UniRef100_A6QQB2 TBC1D10C protein n=1 Tax=Bos taurus RepID=A6QQB... 58 7e-07
UniRef100_A7SHD4 Predicted protein (Fragment) n=1 Tax=Nematostel... 58 7e-07
UniRef100_Q5RCW6 RAB GTPase-activating protein 1-like n=1 Tax=Po... 58 7e-07
UniRef100_UPI000194D829 PREDICTED: RAB GTPase activating protein... 57 9e-07
UniRef100_UPI000186E0D3 conserved hypothetical protein n=1 Tax=P... 57 9e-07
UniRef100_UPI00017EFAAB PREDICTED: similar to Rab GTPase-activat... 57 9e-07
UniRef100_UPI000175FD50 PREDICTED: similar to Rab GTPase-activat... 57 9e-07
UniRef100_UPI00015603CF PREDICTED: similar to Rab GTPase-activat... 57 9e-07
UniRef100_UPI000155E3D4 PREDICTED: similar to MGC130926 protein ... 57 9e-07
UniRef100_UPI000155C9D0 PREDICTED: similar to MGC52980 protein n... 57 9e-07
UniRef100_UPI0000F2BC34 PREDICTED: similar to Coronin, actin bin... 57 9e-07
UniRef100_UPI0000F2B53D PREDICTED: hypothetical protein n=1 Tax=... 57 9e-07
UniRef100_UPI0000D99ECB PREDICTED: similar to RAB GTPase activat... 57 9e-07
UniRef100_UPI0000D99ECA PREDICTED: similar to RAB GTPase activat... 57 9e-07
UniRef100_UPI00005A1360 PREDICTED: similar to RAB GTPase activat... 57 9e-07
UniRef100_UPI00005A135F PREDICTED: similar to RAB GTPase activat... 57 9e-07
UniRef100_UPI0001A2BF61 UPI0001A2BF61 related cluster n=1 Tax=Da... 57 9e-07
UniRef100_UPI0000DA3A76 RAB GTPase activating protein 1-like n=1... 57 9e-07
UniRef100_UPI00016E2DDF UPI00016E2DDF related cluster n=1 Tax=Ta... 57 9e-07
UniRef100_UPI0000EB3702 UPI0000EB3702 related cluster n=1 Tax=Ca... 57 9e-07
UniRef100_UPI000195139C UPI000195139C related cluster n=1 Tax=Bo... 57 9e-07
UniRef100_UPI000179E8C2 Hypothetical protein n=1 Tax=Bos taurus ... 57 9e-07
UniRef100_UPI0000ECA67A RAB GTPase activating protein 1 n=2 Tax=... 57 9e-07
UniRef100_B7Z2B4 cDNA FLJ53389, highly similar to Homo sapiens R... 57 9e-07
UniRef100_A8PWI6 Putative uncharacterized protein n=1 Tax=Malass... 57 9e-07
UniRef100_A5DY91 Putative uncharacterized protein n=1 Tax=Lodder... 57 9e-07
UniRef100_Q8C9V1-3 Isoform 3 of Carabin n=1 Tax=Mus musculus Rep... 57 9e-07
UniRef100_Q8C9V1 Carabin n=2 Tax=Euarchontoglires RepID=TB10C_MOUSE 57 9e-07
UniRef100_Q5RAN1 Rab GTPase-activating protein 1 n=1 Tax=Pongo a... 57 9e-07
UniRef100_Q9Y3P9 Rab GTPase-activating protein 1 n=3 Tax=Hominin... 57 9e-07
UniRef100_A6H6A9 RAB GTPase-activating protein 1-like n=1 Tax=Mu... 57 9e-07
UniRef100_UPI00005066E8 RAB GTPase activating protein 1 n=1 Tax=... 57 1e-06
UniRef100_Q9VSI2 CG7112, isoform A n=1 Tax=Drosophila melanogast... 57 1e-06
UniRef100_Q8MT33 RE63030p n=1 Tax=Drosophila melanogaster RepID=... 57 1e-06
UniRef100_Q2LYM4 GA20112 n=1 Tax=Drosophila pseudoobscura pseudo... 57 1e-06
UniRef100_B4QLN1 GD14085 n=1 Tax=Drosophila simulans RepID=B4QLN... 57 1e-06
UniRef100_B4PG39 GE20734 n=1 Tax=Drosophila yakuba RepID=B4PG39_... 57 1e-06
UniRef100_B4N6D5 GK17754 n=1 Tax=Drosophila willistoni RepID=B4N... 57 1e-06
UniRef100_B4LDR3 GJ12967 n=1 Tax=Drosophila virilis RepID=B4LDR3... 57 1e-06
UniRef100_B4KYD0 GI12826 n=1 Tax=Drosophila mojavensis RepID=B4K... 57 1e-06
UniRef100_B4HJH3 GM25048 n=1 Tax=Drosophila sechellia RepID=B4HJ... 57 1e-06
UniRef100_B3NFT2 GG14306 n=1 Tax=Drosophila erecta RepID=B3NFT2_... 57 1e-06
UniRef100_B3M5J8 GF24391 n=1 Tax=Drosophila ananassae RepID=B3M5... 57 1e-06
UniRef100_A8JNN5 CG7112, isoform B n=1 Tax=Drosophila melanogast... 57 1e-06
UniRef100_A2RVI2 IP18148p (Fragment) n=2 Tax=Drosophila melanoga... 57 1e-06
UniRef100_C7YI07 Putative uncharacterized protein n=1 Tax=Nectri... 57 1e-06
UniRef100_B2W896 GTPase-activating protein GYP5 n=1 Tax=Pyrenoph... 57 1e-06
UniRef100_A7EZE5 Putative uncharacterized protein n=1 Tax=Sclero... 57 1e-06
UniRef100_A6RU09 Putative uncharacterized protein n=1 Tax=Botryo... 57 1e-06
UniRef100_A2AWA9-2 Isoform 2 of Rab GTPase-activating protein 1 ... 57 1e-06
UniRef100_A2AWA9 Rab GTPase-activating protein 1 n=2 Tax=Mus mus... 57 1e-06
UniRef100_Q7ZYI2 MGC52980 protein n=1 Tax=Xenopus laevis RepID=Q... 57 2e-06
UniRef100_B4H627 GL24699 n=1 Tax=Drosophila persimilis RepID=B4H... 57 2e-06
UniRef100_B3RPD6 Putative uncharacterized protein n=1 Tax=Tricho... 57 2e-06
UniRef100_C1G3W9 Putative uncharacterized protein n=1 Tax=Paraco... 57 2e-06
UniRef100_C0S556 Putative uncharacterized protein n=1 Tax=Paraco... 57 2e-06
UniRef100_A8N5V4 Putative uncharacterized protein n=1 Tax=Coprin... 57 2e-06
UniRef100_UPI00017EFC75 PREDICTED: similar to hCG2024869 n=1 Tax... 56 2e-06
UniRef100_UPI000155D36F PREDICTED: similar to RAB GTPase activat... 56 2e-06
UniRef100_Q7PML2 AGAP003924-PA n=1 Tax=Anopheles gambiae RepID=Q... 56 2e-06
UniRef100_A7SQA7 Predicted protein n=1 Tax=Nematostella vectensi... 56 2e-06
UniRef100_Q2UJM3 Rab6 GTPase activator GAPCenA and related TBC d... 56 2e-06
UniRef100_C6HIN6 GTPase activating protein n=1 Tax=Ajellomyces c... 56 2e-06
UniRef100_C5JM07 GTPase activating protein n=1 Tax=Ajellomyces d... 56 2e-06
UniRef100_C5GV36 GTPase activating protein n=1 Tax=Ajellomyces d... 56 2e-06
UniRef100_C5FUP0 GTPase-activating protein GYP5 n=1 Tax=Microspo... 56 2e-06
UniRef100_C1GVR9 Putative uncharacterized protein n=1 Tax=Paraco... 56 2e-06
UniRef100_C0NJZ9 GTPase activating protein GYP5 n=1 Tax=Ajellomy... 56 2e-06
UniRef100_B8N0Y0 GTPase activating protein (Gyp5), putative n=1 ... 56 2e-06
UniRef100_A6QWC7 Putative uncharacterized protein n=1 Tax=Ajello... 56 2e-06
UniRef100_UPI000186DDBC conserved hypothetical protein n=1 Tax=P... 56 3e-06
UniRef100_Q4DHN9 Rab6 GTPase activating protein, putative n=1 Ta... 56 3e-06
UniRef100_B4IZF4 GH15059 n=1 Tax=Drosophila grimshawi RepID=B4IZ... 56 3e-06
UniRef100_B3L8X2 GTPase activator protein, putative n=1 Tax=Plas... 56 3e-06
UniRef100_Q9P4Y9 Putative uncharacterized protein B11B22.030 n=1... 56 3e-06
UniRef100_Q7RUP0 Putative uncharacterized protein n=1 Tax=Neuros... 56 3e-06
UniRef100_Q4P6P1 Putative uncharacterized protein n=1 Tax=Ustila... 56 3e-06
UniRef100_Q0CQM8 Putative uncharacterized protein n=1 Tax=Asperg... 56 3e-06
UniRef100_B0D0J7 Predicted protein n=1 Tax=Laccaria bicolor S238... 56 3e-06
UniRef100_Q8C9V1-2 Isoform 2 of Carabin n=1 Tax=Mus musculus Rep... 56 3e-06
UniRef100_UPI000180C68C PREDICTED: similar to G protein-coupled ... 55 3e-06
UniRef100_UPI000180B056 PREDICTED: similar to HrPET-1, partial n... 55 3e-06
UniRef100_UPI000023DAC0 hypothetical protein FG00632.1 n=1 Tax=G... 55 3e-06
UniRef100_B0S5K6 Novel protein similar to H.sapiens TBC1D10A, TB... 55 3e-06
UniRef100_Q4P8R3 Putative uncharacterized protein n=1 Tax=Ustila... 55 3e-06
UniRef100_C8VL54 GTPase activating protein (Gyp5), putative (AFU... 55 3e-06
UniRef100_B8LWQ5 GTPase activating protein (Gyp5), putative n=1 ... 55 3e-06
UniRef100_B6QQ57 GTPase activating protein (Gyp5), putative n=1 ... 55 3e-06
UniRef100_B6QQ56 GTPase activating protein (Gyp5), putative n=1 ... 55 3e-06
UniRef100_UPI000151B9E2 hypothetical protein PGUG_04833 n=1 Tax=... 55 4e-06
UniRef100_UPI00003BE071 hypothetical protein DEHA0F03696g n=1 Ta... 55 4e-06
UniRef100_C1LZP8 Ecotropic viral integration site, putative n=1 ... 55 4e-06
UniRef100_Q5A5Q9 Putative uncharacterized protein n=1 Tax=Candid... 55 4e-06
UniRef100_C4YKW4 Putative uncharacterized protein n=1 Tax=Candid... 55 4e-06
UniRef100_C4JRP2 Putative uncharacterized protein n=1 Tax=Uncino... 55 4e-06
UniRef100_B5RU86 DEHA2F03322p n=1 Tax=Debaryomyces hansenii RepI... 55 4e-06
UniRef100_Q5R372-5 Isoform 5 of RAB GTPase-activating protein 1-... 55 4e-06
UniRef100_UPI000186E533 rab6 GTPase activating protein, gapcena,... 55 6e-06
UniRef100_Q16XR6 Ecotropic viral integration site n=1 Tax=Aedes ... 55 6e-06
UniRef100_B4IK15 GM13069 n=1 Tax=Drosophila sechellia RepID=B4IK... 55 6e-06
UniRef100_Q1E7J8 Putative uncharacterized protein n=1 Tax=Coccid... 55 6e-06
UniRef100_C5PGF4 TBC domain containing protein n=1 Tax=Coccidioi... 55 6e-06
UniRef100_B9WCY0 GTPase-activating protein, putative n=1 Tax=Can... 55 6e-06
UniRef100_B6JZH2 GTPase-activating protein GYP5 n=1 Tax=Schizosa... 55 6e-06
UniRef100_UPI00019262FA PREDICTED: similar to TBC1 domain family... 54 7e-06
UniRef100_UPI0001866125 hypothetical protein BRAFLDRAFT_90848 n=... 54 7e-06
UniRef100_Q9GPS2 Putative uncharacterized protein n=1 Tax=Dictyo... 54 7e-06
UniRef100_Q550W0 RabGAP/TBC domain-containing protein n=1 Tax=Di... 54 7e-06
UniRef100_C3YCP3 Putative uncharacterized protein n=1 Tax=Branch... 54 7e-06
UniRef100_B9Q845 Ecotropic viral integration site, putative n=1 ... 54 7e-06
UniRef100_B9PMT5 Ecotropic viral integration site, putative n=1 ... 54 7e-06
UniRef100_B6KER5 TBC domain-containing protein n=1 Tax=Toxoplasm... 54 7e-06
UniRef100_B3RNJ2 Putative uncharacterized protein (Fragment) n=1... 54 7e-06
UniRef100_C5M576 Putative uncharacterized protein n=1 Tax=Candid... 54 7e-06
UniRef100_B6HQK2 Pc22g07150 protein n=1 Tax=Penicillium chrysoge... 54 7e-06
UniRef100_UPI0000221A06 hypothetical protein CBG15140 n=1 Tax=Ca... 54 1e-05
UniRef100_B0WKL8 Ecotropic viral integration site n=1 Tax=Culex ... 54 1e-05
UniRef100_A8XLV0 Putative uncharacterized protein n=1 Tax=Caenor... 54 1e-05
UniRef100_C5E272 KLTH0H02640p n=1 Tax=Lachancea thermotolerans C... 54 1e-05
UniRef100_B0CNH8 Predicted protein (Fragment) n=1 Tax=Laccaria b... 54 1e-05