[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
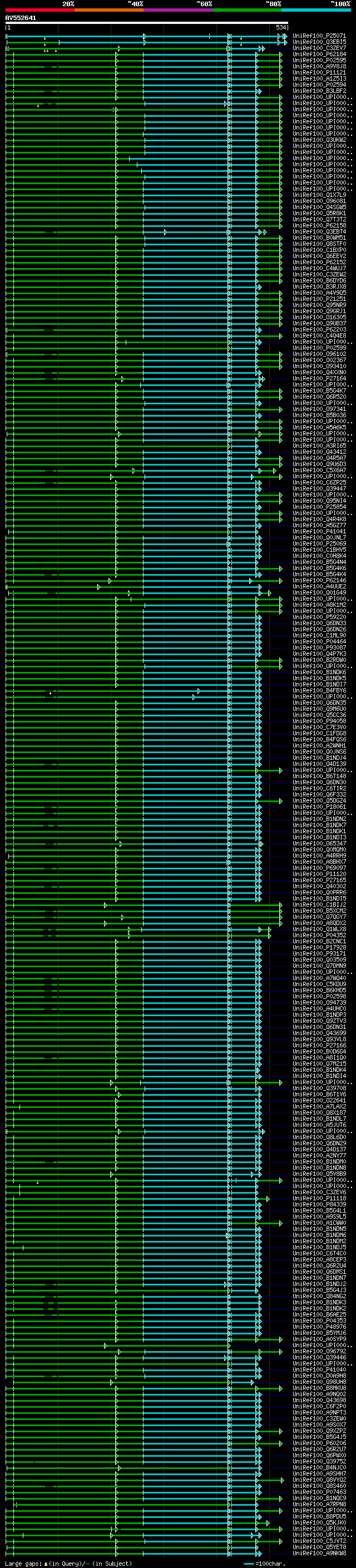
Query= AV552641 RZ35b01R
(534 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_P25071 Calmodulin-like protein 12 n=2 Tax=Arabidopsis ... 289 1e-93
UniRef100_Q3EBI5 Putative uncharacterized protein At2g41100.2 n=... 169 5e-44
UniRef100_C3ZEV7 Putative uncharacterized protein n=1 Tax=Branch... 153 3e-40
UniRef100_P62184 Calmodulin n=1 Tax=Renilla reniformis RepID=CAL... 145 8e-35
UniRef100_P02595 Calmodulin n=1 Tax=Patinopecten sp. RepID=CALM_... 145 8e-35
UniRef100_A9V8J8 Predicted protein n=1 Tax=Monosiga brevicollis ... 144 1e-34
UniRef100_P11121 Calmodulin n=1 Tax=Pyuridae gen. sp. RepID=CALM... 144 3e-34
UniRef100_A1Z5I3 Calmodulin 1b n=1 Tax=Branchiostoma belcheri ts... 142 3e-34
UniRef100_P02594 Calmodulin n=1 Tax=Electrophorus electricus Rep... 143 4e-34
UniRef100_B3LBF2 Calmodulin, putative n=2 Tax=Plasmodium (Plasmo... 142 5e-34
UniRef100_UPI00006179B5 calmodulin-like n=1 Tax=Bos taurus RepID... 141 5e-34
UniRef100_UPI0000E481F6 PREDICTED: similar to calmodulin 2 n=1 T... 142 8e-34
UniRef100_UPI0000E2527E PREDICTED: similar to calmodulin n=1 Tax... 142 8e-34
UniRef100_UPI0001796856 PREDICTED: similar to calmodulin 2 n=1 T... 142 8e-34
UniRef100_UPI0000F2D2EF PREDICTED: hypothetical protein n=1 Tax=... 142 8e-34
UniRef100_UPI0000D9BD62 PREDICTED: similar to calmodulin 1 n=1 T... 142 8e-34
UniRef100_UPI0000EB2E89 Calmodulin (CaM). n=1 Tax=Canis lupus fa... 142 8e-34
UniRef100_Q3UKW2 Putative uncharacterized protein n=1 Tax=Mus mu... 142 8e-34
UniRef100_UPI0001B7AF2E Calmodulin (CaM). n=1 Tax=Rattus norvegi... 142 8e-34
UniRef100_UPI00018815D7 UPI00018815D7 related cluster n=1 Tax=Ho... 142 8e-34
UniRef100_UPI0000D9EC9D PREDICTED: similar to calmodulin 1 isofo... 142 9e-34
UniRef100_UPI0000D9D3FF PREDICTED: similar to calmodulin 1 isofo... 142 9e-34
UniRef100_UPI0000F2B1B4 PREDICTED: hypothetical protein n=1 Tax=... 142 9e-34
UniRef100_UPI0001760975 PREDICTED: similar to calmodulin 2 n=1 T... 142 9e-34
UniRef100_UPI0000E49F67 PREDICTED: similar to calmodulin 2 n=2 T... 142 9e-34
UniRef100_UPI0000E481F7 PREDICTED: similar to calmodulin 2 n=1 T... 142 9e-34
UniRef100_Q1X7L9 Calmodulin 2 n=1 Tax=Apostichopus japonicus Rep... 142 9e-34
UniRef100_O96081 Calmodulin-B n=1 Tax=Halocynthia roretzi RepID=... 142 9e-34
UniRef100_Q4SGW5 Chromosome 14 SCAF14590, whole genome shotgun s... 142 9e-34
UniRef100_Q5R8K1 Putative uncharacterized protein DKFZp469L1534 ... 142 9e-34
UniRef100_Q7T3T2 Calmodulin n=1 Tax=Epinephelus akaara RepID=CAL... 142 9e-34
UniRef100_P62158 Calmodulin n=32 Tax=Gnathostomata RepID=CALM_HUMAN 142 9e-34
UniRef100_Q3EBT4 Putative uncharacterized protein At2g27030.3 n=... 139 1e-33
UniRef100_B0WM51 Calmodulin n=1 Tax=Culex quinquefasciatus RepID... 142 1e-33
UniRef100_Q8STF0 Calmodulin n=1 Tax=Strongylocentrotus intermedi... 142 1e-33
UniRef100_C1BXP0 Calmodulin n=1 Tax=Esox lucius RepID=C1BXP0_ESOLU 142 1e-33
UniRef100_Q6EEV2 Calmodulin n=1 Tax=Pinctada fucata RepID=Q6EEV2... 142 1e-33
UniRef100_P62152 Calmodulin n=28 Tax=Coelomata RepID=CALM_DROME 142 1e-33
UniRef100_C4WUJ7 ACYPI000056 protein n=2 Tax=Neoptera RepID=C4WU... 142 1e-33
UniRef100_C3ZEW2 Putative uncharacterized protein n=1 Tax=Branch... 142 1e-33
UniRef100_B6DYD6 Calmodulin n=1 Tax=Procambarus clarkii RepID=B6... 142 1e-33
UniRef100_B3RJX8 Calmodulin n=1 Tax=Trichoplax adhaerens RepID=B... 142 1e-33
UniRef100_A4V9Q5 Calmodulin-like protein 1 (CaM1) n=2 Tax=Digene... 142 1e-33
UniRef100_P21251 Calmodulin n=1 Tax=Apostichopus japonicus RepID... 142 1e-33
UniRef100_Q95NR9 Calmodulin n=5 Tax=Eumetazoa RepID=CALM_METSE 142 1e-33
UniRef100_Q9GRJ1 Calmodulin n=1 Tax=Lumbricus rubellus RepID=CAL... 142 1e-33
UniRef100_O16305 Calmodulin n=3 Tax=Bilateria RepID=CALM_CAEEL 142 1e-33
UniRef100_Q9UB37 Calmodulin-2 n=1 Tax=Branchiostoma lanceolatum ... 142 1e-33
UniRef100_P62203 Calmodulin n=2 Tax=Plasmodium falciparum RepID=... 141 1e-33
UniRef100_C4Q4E8 Calmodulin, putative n=1 Tax=Schistosoma manson... 141 1e-33
UniRef100_UPI0000E49362 PREDICTED: similar to Calmodulin (CaM) n... 140 1e-33
UniRef100_P02599 Calmodulin n=1 Tax=Dictyostelium discoideum Rep... 140 1e-33
UniRef100_O96102 Calmodulin n=1 Tax=Physarum polycephalum RepID=... 142 1e-33
UniRef100_O02367 Calmodulin n=1 Tax=Ciona intestinalis RepID=CAL... 142 1e-33
UniRef100_O93410 Calmodulin n=1 Tax=Gallus gallus RepID=O93410_C... 141 1e-33
UniRef100_Q4XXN0 Calmodulin, putative n=3 Tax=Plasmodium (Vincke... 141 1e-33
UniRef100_P27164 Calmodulin-related protein n=1 Tax=Petunia x hy... 139 2e-33
UniRef100_UPI0001926FEC PREDICTED: similar to calmodulin n=1 Tax... 140 2e-33
UniRef100_B5G4K7 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 140 2e-33
UniRef100_Q6R520 Calmodulin n=1 Tax=Oreochromis mossambicus RepI... 140 2e-33
UniRef100_UPI00001AA83A PROTEIN (CALMODULIN) n=1 Tax=Escherichia... 141 2e-33
UniRef100_O97341 Calmodulin n=1 Tax=Suberites domuncula RepID=CA... 142 2e-33
UniRef100_B5B036 TCH n=1 Tax=Ipomoea batatas RepID=B5B036_IPOBA 141 2e-33
UniRef100_UPI00017B2E57 UPI00017B2E57 related cluster n=1 Tax=Te... 140 2e-33
UniRef100_A5A6K5 Calmodulin 1 n=1 Tax=Pan troglodytes verus RepI... 140 2e-33
UniRef100_UPI0000587255 PREDICTED: similar to calmodulin B n=1 T... 139 2e-33
UniRef100_UPI0000F2C33A PREDICTED: hypothetical protein n=1 Tax=... 139 3e-33
UniRef100_A3RI65 Calmodulin n=1 Tax=Cicer arietinum RepID=A3RI65... 139 3e-33
UniRef100_Q43412 Calmodulin n=1 Tax=Bidens pilosa RepID=Q43412_B... 140 3e-33
UniRef100_Q4R5A7 Brain cDNA, clone: QtrA-13982, similar to human... 140 3e-33
UniRef100_Q9U6D3 Calmodulin n=1 Tax=Myxine glutinosa RepID=CALM_... 140 3e-33
UniRef100_C5X6A7 Putative uncharacterized protein Sb02g043510 n=... 139 4e-33
UniRef100_UPI000186E8F7 calmodulin-A n=1 Tax=Pediculus humanus c... 142 4e-33
UniRef100_C6ZP25 Calmodulin 1 n=1 Tax=Capsicum annuum RepID=C6ZP... 140 4e-33
UniRef100_Q39447 Calmodulin-2 n=1 Tax=Capsicum annuum RepID=Q394... 140 4e-33
UniRef100_UPI000180B772 PREDICTED: similar to Calmodulin CG8472-... 140 4e-33
UniRef100_Q95NI4 Calmodulin n=1 Tax=Halichondria okadai RepID=CA... 140 4e-33
UniRef100_P25854 Calmodulin-1/4 n=1 Tax=Arabidopsis thaliana Rep... 140 4e-33
UniRef100_UPI00015FF4E8 calmodulin 1 (phosphorylase kinase, delt... 139 4e-33
UniRef100_Q4R4K8 Brain cDNA, clone: QnpA-15172, similar to human... 139 4e-33
UniRef100_A5GZ77 Calmodulin n=2 Tax=Aegiceras corniculatum RepID... 140 5e-33
UniRef100_P41041 Calmodulin n=1 Tax=Pneumocystis carinii RepID=C... 137 5e-33
UniRef100_Q0JNL7 Calmodulin-3 n=2 Tax=Oryza sativa Japonica Grou... 140 5e-33
UniRef100_P25069 Calmodulin-2/3/5 n=4 Tax=Brassicaceae RepID=CAL... 139 5e-33
UniRef100_C1BHV5 Calmodulin n=1 Tax=Oncorhynchus mykiss RepID=C1... 139 5e-33
UniRef100_C0H8K4 Calmodulin n=1 Tax=Salmo salar RepID=C0H8K4_SALSA 139 5e-33
UniRef100_B5G4N4 Putative calmodulin variant 3 n=1 Tax=Taeniopyg... 139 5e-33
UniRef100_B5G4K6 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 139 5e-33
UniRef100_B5G4K4 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 139 5e-33
UniRef100_P62146 Calmodulin-alpha (Fragment) n=3 Tax=Deuterostom... 142 5e-33
UniRef100_A4UUE2 Calmodulin (Fragment) n=1 Tax=Hyriopsis cumingi... 143 7e-33
UniRef100_Q01G49 Calmodulin mutant SYNCAM9 (ISS) n=1 Tax=Ostreoc... 138 7e-33
UniRef100_UPI00017C33EC PREDICTED: similar to calmodulin 2 n=1 T... 142 7e-33
UniRef100_A8K1M2 cDNA FLJ75174, highly similar to Homo sapiens c... 139 7e-33
UniRef100_UPI00005C066E PREDICTED: similar to calmodulin 2 n=1 T... 142 7e-33
UniRef100_P59220 Calmodulin-7 n=30 Tax=Magnoliophyta RepID=CALM7... 139 7e-33
UniRef100_Q6DN33 Calmodulin cam-203 n=1 Tax=Daucus carota RepID=... 139 7e-33
UniRef100_Q6DN26 Calmodulin cam-210 n=1 Tax=Daucus carota RepID=... 139 7e-33
UniRef100_C1ML90 Calmodulin n=1 Tax=Micromonas pusilla CCMP1545 ... 139 7e-33
UniRef100_P04464 Calmodulin n=1 Tax=Triticum aestivum RepID=CALM... 139 7e-33
UniRef100_P93087 Calmodulin n=8 Tax=core eudicotyledons RepID=CA... 139 7e-33
UniRef100_Q4P7K3 CLM_PLEOS Calmodulin (CaM) n=1 Tax=Ustilago may... 139 7e-33
UniRef100_B2RDW0 cDNA, FLJ96792, highly similar to Homo sapiens ... 139 7e-33
UniRef100_UPI000179E504 UPI000179E504 related cluster n=1 Tax=Bo... 142 7e-33
UniRef100_B1NDK6 Calmodulin n=1 Tax=Actinidia kolomikta RepID=B1... 139 7e-33
UniRef100_B1NDK5 Calmodulin n=4 Tax=Actinidia RepID=B1NDK5_9ERIC 139 7e-33
UniRef100_B1NDI7 Calmodulin n=1 Tax=Actinidia deliciosa var. chl... 139 7e-33
UniRef100_B4FBY6 Putative uncharacterized protein n=1 Tax=Zea ma... 139 9e-33
UniRef100_UPI0000DD8A7A Os01g0267900 n=1 Tax=Oryza sativa Japoni... 139 9e-33
UniRef100_Q6DN35 Calmodulin cam-201 n=1 Tax=Daucus carota RepID=... 139 9e-33
UniRef100_Q9M6U0 Calmodulin n=1 Tax=Brassica napus RepID=Q9M6U0_... 139 9e-33
UniRef100_Q5CC36 Calmodulin n=1 Tax=Quercus petraea RepID=Q5CC36... 139 9e-33
UniRef100_P94058 Calmodulin TaCaM2-2 n=1 Tax=Triticum aestivum R... 139 9e-33
UniRef100_C7E3V0 Calmodulin n=1 Tax=Saccharum officinarum RepID=... 139 9e-33
UniRef100_C1FDG8 Calmodulin n=1 Tax=Micromonas sp. RCC299 RepID=... 139 9e-33
UniRef100_B4FQS6 Putative uncharacterized protein n=1 Tax=Zea ma... 139 9e-33
UniRef100_A2WNH1 Calmodulin-3 n=2 Tax=Oryza sativa Indica Group ... 139 9e-33
UniRef100_Q0JNS6 Calmodulin-1 n=14 Tax=Magnoliophyta RepID=CALM1... 139 9e-33
UniRef100_B1NDJ4 Calmodulin n=3 Tax=Actinidiaceae RepID=B1NDJ4_9... 139 9e-33
UniRef100_Q4D139 Calmodulin, putative n=1 Tax=Trypanosoma cruzi ... 138 1e-32
UniRef100_UPI0000D9448E PREDICTED: similar to calmodulin n=1 Tax... 140 1e-32
UniRef100_B6T148 Calmodulin n=1 Tax=Zea mays RepID=B6T148_MAIZE 139 1e-32
UniRef100_Q6DN30 Calmodulin cam-206 n=1 Tax=Daucus carota RepID=... 139 1e-32
UniRef100_C6TIR2 Putative uncharacterized protein n=1 Tax=Glycin... 139 1e-32
UniRef100_Q6F332 Calmodulin-2 n=3 Tax=Oryza sativa RepID=CALM2_O... 139 1e-32
UniRef100_Q5DGZ4 Putative uncharacterized protein n=1 Tax=Schist... 138 1e-32
UniRef100_P18061 Calmodulin n=6 Tax=Trypanosomatidae RepID=CALM_... 138 1e-32
UniRef100_UPI0000182578 PREDICTED: similar to calmodulin 1 n=1 T... 138 1e-32
UniRef100_B1NDN2 Calmodulin n=1 Tax=Actinidia polygama RepID=B1N... 139 1e-32
UniRef100_B1NDK7 Calmodulin n=1 Tax=Actinidia sabiifolia RepID=B... 139 1e-32
UniRef100_B1NDK1 Calmodulin n=1 Tax=Clematoclethra scandens subs... 139 1e-32
UniRef100_B1NDI3 Calmodulin n=14 Tax=core eudicotyledons RepID=B... 139 1e-32
UniRef100_O65347 Calmodulin n=1 Tax=Apium graveolens RepID=O6534... 137 2e-32
UniRef100_Q0MQM0 Calmodulin n=1 Tax=Betula halophila RepID=Q0MQM... 138 2e-32
UniRef100_A4RRH9 Predicted protein n=1 Tax=Ostreococcus lucimari... 138 2e-32
UniRef100_A8BHX7 Calmodulin n=1 Tax=Noccaea caerulescens RepID=A... 138 2e-32
UniRef100_P69097 Calmodulin n=4 Tax=Trypanosoma brucei RepID=CAL... 138 2e-32
UniRef100_P11120 Calmodulin n=1 Tax=Pleurotus cornucopiae RepID=... 137 2e-32
UniRef100_P27165 Calmodulin n=3 Tax=Oomycetes RepID=CALM_PHYIN 137 2e-32
UniRef100_Q40302 Calmodulin n=1 Tax=Macrocystis pyrifera RepID=C... 137 2e-32
UniRef100_Q0PRR6 Calmodulin (Fragment) n=1 Tax=Vigna radiata var... 138 2e-32
UniRef100_B1NDI5 Calmodulin n=1 Tax=Actinidia chinensis RepID=B1... 138 2e-32
UniRef100_C1BIJ2 Calmodulin-alpha n=1 Tax=Osmerus mordax RepID=C... 142 2e-32
UniRef100_B5XCM2 Calmodulin n=1 Tax=Salmo salar RepID=B5XCM2_SALSA 142 2e-32
UniRef100_Q7QGY7 AGAP010957-PA (Fragment) n=1 Tax=Anopheles gamb... 142 2e-32
UniRef100_A8QDX2 Calmodulin, putative n=1 Tax=Brugia malayi RepI... 142 2e-32
UniRef100_Q1WLX8 Calmodulin n=1 Tax=Chlamydomonas incerta RepID=... 137 2e-32
UniRef100_P04352 Calmodulin n=2 Tax=Chlamydomonas reinhardtii Re... 137 2e-32
UniRef100_B2CNC1 Calmodulin n=1 Tax=Beta vulgaris RepID=B2CNC1_B... 138 2e-32
UniRef100_P17928 Calmodulin n=5 Tax=Papilionoideae RepID=CALM_MEDSA 138 2e-32
UniRef100_P93171 Calmodulin n=2 Tax=core eudicotyledons RepID=CA... 138 2e-32
UniRef100_Q03509 Calmodulin-6 n=1 Tax=Arabidopsis thaliana RepID... 138 2e-32
UniRef100_Q7DMN9 Calmodulin-5/6/7/8 n=3 Tax=Solanum RepID=CALM5_... 138 2e-32
UniRef100_UPI000186176F hypothetical protein BRAFLDRAFT_120113 n... 137 2e-32
UniRef100_A7WQ40 Calmodulin n=1 Tax=Noctiluca scintillans RepID=... 137 2e-32
UniRef100_C5KDU9 Calmodulin, putative n=1 Tax=Perkinsus marinus ... 137 2e-32
UniRef100_B6KHD5 Calmodulin n=4 Tax=Toxoplasma gondii RepID=B6KH... 137 2e-32
UniRef100_P02598 Calmodulin n=2 Tax=Tetrahymena RepID=CALM_TETPY 137 2e-32
UniRef100_O94739 Calmodulin n=1 Tax=Pleurotus ostreatus RepID=CA... 137 2e-32
UniRef100_A4UHC0 Calmodulin n=4 Tax=Dinophyceae RepID=CALM_ALEFU 137 2e-32
UniRef100_B1NDP3 Calmodulin n=1 Tax=Actinidia kolomikta RepID=B1... 139 2e-32
UniRef100_Q9ZTV3 Calmodulin n=1 Tax=Phaseolus vulgaris RepID=Q9Z... 137 3e-32
UniRef100_Q6DN31 Calmodulin cam-205 n=1 Tax=Daucus carota RepID=... 137 3e-32
UniRef100_Q43699 Putative uncharacterized protein n=1 Tax=Zea ma... 137 3e-32
UniRef100_Q93VL8 Calmodulin n=3 Tax=Magnoliophyta RepID=Q93VL8_P... 137 3e-32
UniRef100_P27166 Calmodulin n=1 Tax=Stylonychia lemnae RepID=CAL... 137 3e-32
UniRef100_B0D6G4 Predicted protein n=2 Tax=Eukaryota RepID=B0D6G... 137 3e-32
UniRef100_A8I1Q0 Calmodulin n=1 Tax=Heterocapsa triquetra RepID=... 137 3e-32
UniRef100_Q7M215 Calmodulin n=1 Tax=Pisum sativum RepID=Q7M215_PEA 138 3e-32
UniRef100_B1NDK4 Calmodulin n=1 Tax=Actinidia deliciosa var. chl... 137 3e-32
UniRef100_B1NDI4 Calmodulin n=1 Tax=Actinidia chinensis RepID=B1... 137 3e-32
UniRef100_UPI0000F2C33B PREDICTED: similar to calmodulin 2 n=1 T... 135 3e-32
UniRef100_Q39708 Calmodulin-like protein n=1 Tax=Dunaliella sali... 139 3e-32
UniRef100_B6T1V6 Calmodulin n=1 Tax=Zea mays RepID=B6T1V6_MAIZE 137 3e-32
UniRef100_O22641 Calmodulin n=1 Tax=Zea mays RepID=O22641_MAIZE 137 3e-32
UniRef100_A7LAX2 Calmodulin 1 n=1 Tax=Morus nigra RepID=A7LAX2_M... 137 3e-32
UniRef100_Q8X187 Calmodulin n=2 Tax=Paxillus involutus RepID=CAL... 137 3e-32
UniRef100_B1NDL7 Calmodulin n=1 Tax=Actinidia deliciosa var. chl... 138 3e-32
UniRef100_A5JUT6 Calmodulin n=2 Tax=Magnoliophyta RepID=A5JUT6_W... 137 3e-32
UniRef100_UPI0001861774 hypothetical protein BRAFLDRAFT_209901 n... 137 3e-32
UniRef100_Q8L6D0 Putative calmodulin n=1 Tax=Solanum commersonii... 137 5e-32
UniRef100_Q6DN29 Caomodulin cam-207 n=1 Tax=Daucus carota RepID=... 137 5e-32
UniRef100_Q4D137 Calmodulin n=1 Tax=Trypanosoma cruzi RepID=Q4D1... 136 5e-32
UniRef100_A2NY77 Calmodulin n=1 Tax=Physcomitrella patens RepID=... 135 5e-32
UniRef100_B1NDM0 Calmodulin n=1 Tax=Actinidia deliciosa var. del... 139 5e-32
UniRef100_B1NDN8 Calmodulin n=1 Tax=Actinidia eriantha f. alba R... 137 5e-32
UniRef100_Q5V8B9 Calmodulin (Fragment) n=1 Tax=Paxillus involutu... 138 5e-32
UniRef100_UPI000059FE19 PREDICTED: similar to calmodulin 1 isofo... 135 6e-32
UniRef100_UPI0001861775 hypothetical protein BRAFLDRAFT_277837 n... 134 6e-32
UniRef100_C3ZEV6 Putative uncharacterized protein n=1 Tax=Branch... 134 6e-32
UniRef100_P11118 Calmodulin n=2 Tax=Euglena gracilis RepID=CALM_... 136 6e-32
UniRef100_P84339 Calmodulin n=1 Tax=Agaricus bisporus RepID=CALM... 136 6e-32
UniRef100_B5G4L1 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 135 6e-32
UniRef100_A9S9L5 Predicted protein n=1 Tax=Physcomitrella patens... 134 6e-32
UniRef100_A1CWW0 Calmodulin n=1 Tax=Neosartorya fischeri NRRL 18... 134 6e-32
UniRef100_B1NDN5 Calmodulin n=1 Tax=Actinidia deliciosa var. chl... 136 6e-32
UniRef100_B1NDM6 Calmodulin n=1 Tax=Actinidia melliana RepID=B1N... 136 6e-32
UniRef100_B1NDM2 Calmodulin n=1 Tax=Actinidia valvata RepID=B1ND... 136 6e-32
UniRef100_B1NDJ5 Calmodulin n=1 Tax=Actinidia kolomikta RepID=B1... 136 6e-32
UniRef100_C6T4C0 Putative uncharacterized protein n=1 Tax=Glycin... 136 8e-32
UniRef100_A8CEP3 Calmodulin n=1 Tax=Saccharina japonica RepID=CA... 135 8e-32
UniRef100_Q6R2U4 Calmodulin n=1 Tax=Arachis hypogaea RepID=Q6R2U... 136 8e-32
UniRef100_Q6DMS1 Calmodulin n=1 Tax=Salvia miltiorrhiza RepID=Q6... 136 8e-32
UniRef100_B1NDN7 Calmodulin n=1 Tax=Actinidia eriantha f. alba R... 136 8e-32
UniRef100_B1NDJ2 Calmodulin n=1 Tax=Actinidia sabiifolia RepID=B... 136 8e-32
UniRef100_B5G4J3 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 135 8e-32
UniRef100_Q84NG2 Calmodulin (Fragment) n=1 Tax=Pyrus communis Re... 139 1e-31
UniRef100_B1NDK3 Calmodulin n=1 Tax=Actinidia sabiifolia RepID=B... 139 1e-31
UniRef100_B1NDK2 Calmodulin n=1 Tax=Actinidia sabiifolia RepID=B... 139 1e-31
UniRef100_B6AE25 Calmodulin , putative n=1 Tax=Cryptosporidium m... 137 1e-31
UniRef100_P04353 Calmodulin n=1 Tax=Spinacia oleracea RepID=CALM... 135 1e-31
UniRef100_P48976 Calmodulin n=1 Tax=Malus x domestica RepID=CALM... 135 1e-31
UniRef100_B5YMJ6 Calmodulin n=1 Tax=Thalassiosira pseudonana CCM... 135 1e-31
UniRef100_A0SYP9 Calmodulin n=3 Tax=Sclerotiniaceae RepID=A0SYP9... 134 1e-31
UniRef100_UPI0001923CB0 PREDICTED: similar to Calmodulin CG8472-... 139 1e-31
UniRef100_O96792 Calmodulin-like protein CaML3 n=1 Tax=Branchios... 135 1e-31
UniRef100_Q39446 Calmodulin-1 n=1 Tax=Capsicum annuum RepID=Q394... 135 1e-31
UniRef100_UPI000179E6C6 Similar to calmodulin n=1 Tax=Bos taurus... 134 1e-31
UniRef100_P41040 Calmodulin n=1 Tax=Zea mays RepID=CALM_MAIZE 135 1e-31
UniRef100_D0A9H8 Calmodulin, putative, (Fragment) n=1 Tax=Trypan... 138 1e-31
UniRef100_Q98UH8 Calmodulin (Fragment) n=1 Tax=Clemmys japonica ... 137 1e-31
UniRef100_B8MKU8 Calmodulin n=1 Tax=Talaromyces stipitatus ATCC ... 133 2e-31
UniRef100_A9NQ02 Putative uncharacterized protein n=1 Tax=Picea ... 135 2e-31
UniRef100_Q43698 Calmodulin n=1 Tax=Zea mays RepID=Q43698_MAIZE 135 2e-31
UniRef100_C6F2P0 Putative calmodulin n=4 Tax=Cupressaceae RepID=... 135 2e-31
UniRef100_A9NPT3 Putative uncharacterized protein n=1 Tax=Picea ... 135 2e-31
UniRef100_C3ZEW0 Putative uncharacterized protein n=1 Tax=Branch... 135 2e-31
UniRef100_A9S0X7 Predicted protein n=1 Tax=Physcomitrella patens... 134 2e-31
UniRef100_Q9XZP2 Calmodulin-2 n=1 Tax=Branchiostoma floridae Rep... 134 2e-31
UniRef100_B5G4J5 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 134 2e-31
UniRef100_P60206 Calmodulin n=18 Tax=Eurotiomycetidae RepID=CALM... 133 2e-31
UniRef100_Q6R2U7 Calmodulin n=1 Tax=Arachis hypogaea RepID=Q6R2U... 135 2e-31
UniRef100_Q6PWX0 Calmodulin n=1 Tax=Arachis hypogaea RepID=Q6PWX... 135 2e-31
UniRef100_Q39752 Calmodulin n=1 Tax=Fagus sylvatica RepID=CALM_F... 135 2e-31
UniRef100_B4NJC0 GK18988 n=1 Tax=Drosophila willistoni RepID=B4N... 134 2e-31
UniRef100_A9SHH7 Predicted protein n=1 Tax=Physcomitrella patens... 134 2e-31
UniRef100_Q8VYQ2 Calmodulin n=1 Tax=Vitis vinifera RepID=Q8VYQ2_... 134 3e-31
UniRef100_Q8S460 Calmodulin n=1 Tax=Sonneratia paracaseolaris Re... 134 3e-31
UniRef100_P07463 Calmodulin n=1 Tax=Paramecium tetraurelia RepID... 134 3e-31
UniRef100_B1NQC9 Putative uncharacterized protein n=1 Tax=Stachy... 132 3e-31
UniRef100_A7RPN8 Predicted protein (Fragment) n=1 Tax=Nematostel... 134 3e-31
UniRef100_UPI00004C0EFB PREDICTED: similar to calmodulin 1 n=1 T... 134 4e-31
UniRef100_B8PDU5 Calmodulin n=1 Tax=Postia placenta Mad-698-R Re... 133 4e-31
UniRef100_Q5KJK0 Putative uncharacterized protein n=1 Tax=Filoba... 133 4e-31
UniRef100_UPI0000F2CE9A PREDICTED: similar to calmodulin n=1 Tax... 132 5e-31
UniRef100_UPI0000F2E443 PREDICTED: similar to calmodulin n=1 Tax... 135 5e-31
UniRef100_C5JVT2 Calmodulin A n=1 Tax=Ajellomyces dermatitidis S... 131 5e-31
UniRef100_Q5YET8 Calmodulin n=1 Tax=Bigelowiella natans RepID=Q5... 132 5e-31
UniRef100_A9NKW8 Putative uncharacterized protein n=1 Tax=Picea ... 133 5e-31
UniRef100_O82018 Calmodulin n=1 Tax=Mougeotia scalaris RepID=CAL... 133 5e-31
UniRef100_Q9HFY6 Calmodulin n=1 Tax=Blastocladiella emersonii Re... 133 5e-31
UniRef100_B7GD08 Calmoduline n=1 Tax=Phaeodactylum tricornutum C... 132 5e-31
UniRef100_C3ZEV9 Putative uncharacterized protein n=1 Tax=Branch... 132 5e-31
UniRef100_P61861 Calmodulin n=13 Tax=Pezizomycotina RepID=CALM_C... 131 5e-31
UniRef100_Q9LDQ9 Calmodulin n=1 Tax=Chara corallina RepID=Q9LDQ9... 133 5e-31
UniRef100_Q9BRL5 CALM3 protein n=1 Tax=Homo sapiens RepID=Q9BRL5... 137 5e-31
UniRef100_A8NMQ1 Calmodulin n=1 Tax=Coprinopsis cinerea okayama7... 137 5e-31
UniRef100_P62150 Calmodulin-A (Fragment) n=4 Tax=Euteleostomi Re... 135 5e-31
UniRef100_A9NRI1 Putative uncharacterized protein n=1 Tax=Picea ... 133 6e-31
UniRef100_B1NDN0 Calmodulin n=1 Tax=Actinidia arguta RepID=B1NDN... 134 6e-31
UniRef100_B0D6G1 Predicted protein n=1 Tax=Laccaria bicolor S238... 132 6e-31
UniRef100_UPI0001AE647A UPI0001AE647A related cluster n=1 Tax=Ho... 137 6e-31
UniRef100_A3FQ56 Calmodulin n=2 Tax=Cryptosporidium RepID=A3FQ56... 134 8e-31
UniRef100_UPI0000D92986 PREDICTED: similar to calmodulin n=1 Tax... 134 8e-31
UniRef100_Q29376 Calmodulin (Fragment) n=1 Tax=Sus scrofa RepID=... 136 8e-31
UniRef100_A7Y374 Calmodulin (Fragment) n=1 Tax=Crassostrea gigas... 131 8e-31
UniRef100_Q9D6P8 Calmodulin-like protein 3 n=1 Tax=Mus musculus ... 132 1e-30
UniRef100_P15094 Calmodulin n=1 Tax=Achlya klebsiana RepID=CALM_... 131 1e-30
UniRef100_Q25420 Calmodulin (Fragment) n=1 Tax=Leishmania tarent... 132 1e-30
UniRef100_UPI00018815D8 UPI00018815D8 related cluster n=1 Tax=Ho... 131 1e-30
UniRef100_Q8LRL0 Calmodulin 1 n=1 Tax=Ceratopteris richardii Rep... 132 1e-30
UniRef100_A9RWJ4 Predicted protein n=1 Tax=Physcomitrella patens... 131 1e-30
UniRef100_UPI00005A1895 PREDICTED: similar to calmodulin 1 isofo... 131 1e-30
UniRef100_P53440 Calmodulin, flagellar n=1 Tax=Naegleria gruberi... 133 2e-30
UniRef100_Q1HCM6 Calmodulin n=1 Tax=Phytomonas serpens RepID=Q1H... 131 2e-30
UniRef100_UPI0001861771 hypothetical protein BRAFLDRAFT_120115 n... 130 2e-30
UniRef100_UPI000186176D hypothetical protein BRAFLDRAFT_72666 n=... 131 2e-30
UniRef100_UPI0001555597 PREDICTED: similar to Chain D, Crystal S... 130 2e-30
UniRef100_P05932 Calmodulin-beta (Fragment) n=1 Tax=Arbacia punc... 131 2e-30
UniRef100_UPI000155519E PREDICTED: similar to calmodulin n=1 Tax... 135 2e-30
UniRef100_UPI0001552F4D PREDICTED: similar to calmodulin n=1 Tax... 130 2e-30
UniRef100_UPI0000E222C4 PREDICTED: hypothetical protein n=1 Tax=... 133 2e-30
UniRef100_P27482 Calmodulin-like protein 3 n=1 Tax=Homo sapiens ... 133 2e-30
UniRef100_B9N3A0 Predicted protein n=1 Tax=Populus trichocarpa R... 131 2e-30
UniRef100_A9RNC0 Predicted protein n=1 Tax=Physcomitrella patens... 130 2e-30
UniRef100_Q5C0Z2 SJCHGC00574 protein (Fragment) n=2 Tax=Bilateri... 130 2e-30
UniRef100_UPI00019254ED PREDICTED: similar to calmodulin n=1 Tax... 129 3e-30
UniRef100_Q5U206 Calmodulin-like protein 3 n=1 Tax=Rattus norveg... 130 3e-30
UniRef100_UPI0000ECD0CE Calmodulin, striated muscle. n=2 Tax=Gal... 134 3e-30
UniRef100_Q4T6S4 Chromosome 10 SCAF8630, whole genome shotgun se... 129 4e-30
UniRef100_UPI0001A7B2F8 CAM1 (CALMODULIN 1); calcium ion binding... 130 4e-30
UniRef100_C1BT99 Calmodulin n=1 Tax=Lepeophtheirus salmonis RepI... 130 4e-30
UniRef100_A5BNP0 Putative uncharacterized protein n=1 Tax=Vitis ... 130 4e-30
UniRef100_P13868 Calmodulin-1 n=1 Tax=Solanum tuberosum RepID=CA... 130 4e-30
UniRef100_Q4N4C2 Calmodulin, putative n=1 Tax=Theileria parva Re... 129 4e-30
UniRef100_A9PCR6 Putative uncharacterized protein n=1 Tax=Populu... 130 5e-30
UniRef100_O17501 Calmodulin 2 (Fragment) n=1 Tax=Branchiostoma l... 134 5e-30
UniRef100_Q6BS94 DEHA2D10582p n=2 Tax=Debaryomyces hansenii RepI... 134 5e-30
UniRef100_UPI000059FE1A PREDICTED: similar to calmodulin 1 isofo... 129 7e-30
UniRef100_A9PDT9 Predicted protein n=1 Tax=Populus trichocarpa R... 129 9e-30
UniRef100_Q4UF72 Calmodulin, putative n=1 Tax=Theileria annulata... 128 9e-30
UniRef100_Q32W15 Calmodulin (Fragment) n=2 Tax=Campanulariidae R... 133 9e-30
UniRef100_Q1ALF5 Calmodulin (Fragment) n=29 Tax=Leptomedusae Rep... 133 9e-30
UniRef100_C5IJ81 Calmodulin isoform 1 n=1 Tax=Solanum tuberosum ... 128 1e-29
UniRef100_P27161 Calmodulin n=4 Tax=Solanaceae RepID=CALM_SOLLC 128 1e-29
UniRef100_UPI000194E1BC PREDICTED: calmodulin 3 (phosphorylase k... 132 1e-29
UniRef100_Q32W00 Calmodulin (Fragment) n=1 Tax=Laomedea calceoli... 132 1e-29
UniRef100_Q6R2U6 Calmodulin n=1 Tax=Arachis hypogaea RepID=Q6R2U... 128 2e-29
UniRef100_Q32W01 Calmodulin (Fragment) n=1 Tax=Obelia dichotoma ... 132 2e-29
UniRef100_P02597 Calmodulin, striated muscle n=1 Tax=Gallus gall... 132 2e-29
UniRef100_B6T376 Calmodulin n=1 Tax=Zea mays RepID=B6T376_MAIZE 128 2e-29
UniRef100_Q5MCR7 Calmodulin 2 n=1 Tax=Codonopsis lanceolata RepI... 127 2e-29
UniRef100_A7AWR1 Calmodulin n=1 Tax=Babesia bovis RepID=A7AWR1_B... 127 2e-29
UniRef100_C3ZEV8 Putative uncharacterized protein n=1 Tax=Branch... 132 2e-29
UniRef100_Q6C3K3 YALI0E34111p n=1 Tax=Yarrowia lipolytica RepID=... 127 3e-29
UniRef100_UPI00005002F3 similar to calmodulin 1 (LOC640703), mRN... 131 3e-29
UniRef100_Q32W36 Calmodulin (Fragment) n=1 Tax=Silicularia rosea... 131 3e-29
UniRef100_UPI00017586FD PREDICTED: similar to calmodulin 2 n=1 T... 125 3e-29
UniRef100_Q5CC38 Calmodulin n=1 Tax=Quercus petraea RepID=Q5CC38... 126 3e-29
UniRef100_Q32W33 Calmodulin (Fragment) n=1 Tax=Orthopyxis integr... 131 3e-29
UniRef100_Q32VZ5 Calmodulin (Fragment) n=1 Tax=Eucheilota bakeri... 131 3e-29
UniRef100_Q3LRX2 Calmodulin 1 n=1 Tax=Catharanthus roseus RepID=... 127 4e-29
UniRef100_A8Y7S8 Z-box binding factor 3 n=1 Tax=Arabidopsis thal... 127 4e-29
UniRef100_O15931 Calmodulin (Fragment) n=3 Tax=Dinophyceae RepID... 126 4e-29
UniRef100_Q9M428 Putative calmodulin (Fragment) n=1 Tax=Oryza sa... 128 4e-29
UniRef100_UPI0000ECC853 Calmodulin (CaM). n=1 Tax=Gallus gallus ... 130 5e-29
UniRef100_UPI0000ECBDA1 Neo-calmodulin (NeoCaM) n=1 Tax=Gallus g... 130 5e-29
UniRef100_Q4RN51 Chromosome undetermined SCAF15016, whole genome... 130 5e-29
UniRef100_Q32VZ8 Calmodulin (Fragment) n=1 Tax=Obelia geniculata... 130 5e-29
UniRef100_UPI0001863ED1 hypothetical protein BRAFLDRAFT_114722 n... 123 6e-29
UniRef100_B4JT33 GH23405 n=1 Tax=Drosophila grimshawi RepID=B4JT... 122 6e-29
UniRef100_P27163 Calmodulin-2 n=1 Tax=Petunia x hybrida RepID=CA... 126 6e-29
UniRef100_P05419 Neo-calmodulin (Fragment) n=1 Tax=Gallus gallus... 128 6e-29
UniRef100_Q32W03 Calmodulin (Fragment) n=5 Tax=Campanulariidae R... 130 6e-29
UniRef100_Q32VZ7 Calmodulin (Fragment) n=1 Tax=Obelia geniculata... 130 6e-29
UniRef100_O45209 Calmodulin-like protein (Fragment) n=1 Tax=Bran... 114 7e-29
UniRef100_UPI0001A7B2F7 CAM1 (CALMODULIN 1); calcium ion binding... 125 7e-29
UniRef100_A2DXW5 Calmodulin, putative n=1 Tax=Trichomonas vagina... 127 7e-29
UniRef100_C3YZA8 Putative uncharacterized protein n=1 Tax=Branch... 122 7e-29
UniRef100_B6QN11 Calmodulin, putative n=1 Tax=Penicillium marnef... 117 7e-29
UniRef100_B6K825 Calmodulin Cam1 n=1 Tax=Schizosaccharomyces jap... 130 8e-29
UniRef100_Q40642 Calmodulin-like protein 1 n=1 Tax=Oryza sativa ... 126 9e-29
UniRef100_A9NMR6 Putative uncharacterized protein n=1 Tax=Picea ... 126 1e-28
UniRef100_UPI0001863ED3 hypothetical protein BRAFLDRAFT_123309 n... 124 1e-28
UniRef100_B5DYP2 GA26322 n=2 Tax=pseudoobscura subgroup RepID=B5... 125 1e-28
UniRef100_Q5VIT6 Calmodulin (Fragment) n=2 Tax=Penicillium RepID... 125 1e-28
UniRef100_Q32VZ9 Calmodulin (Fragment) n=1 Tax=Obelia geniculata... 129 1e-28
UniRef100_Q32VZ2 Calmodulin (Fragment) n=1 Tax=Opercularella pum... 129 1e-28
UniRef100_B3G4T9 Calmodulin (Fragment) n=61 Tax=Stylasteridae Re... 129 1e-28
UniRef100_Q59Q76 Calmodulin n=1 Tax=Candida albicans RepID=Q59Q7... 129 1e-28
UniRef100_B9WGP8 Calmodulin, putative (Cam, putative) n=1 Tax=Ca... 129 1e-28
UniRef100_A3GH66 Calmodulin n=1 Tax=Pichia stipitis RepID=A3GH66... 129 1e-28
UniRef100_P05933 Calmodulin n=1 Tax=Schizosaccharomyces pombe Re... 129 1e-28
UniRef100_UPI0000513C25 PREDICTED: similar to Calmodulin CG8472-... 125 1e-28
UniRef100_Q8S1Y9 Calmodulin-like protein 1 n=2 Tax=Oryza sativa ... 125 2e-28
UniRef100_B2BG00 Calmodulin (Fragment) n=166 Tax=Trichocomaceae ... 126 2e-28
UniRef100_UPI000069E4E3 Cmd-1-prov protein. n=1 Tax=Xenopus (Sil... 129 2e-28
UniRef100_Q7SZ95 Cam protein (Fragment) n=1 Tax=Xenopus laevis R... 129 2e-28
UniRef100_C4Q4E7 Calmodulin, putative n=1 Tax=Schistosoma manson... 129 2e-28
UniRef100_B3F7W1 Calmodulin (Fragment) n=5 Tax=Trichocomaceae Re... 126 2e-28
UniRef100_A9XEW6 Calmodulin (Fragment) n=3 Tax=Geosmithia RepID=... 126 2e-28
UniRef100_P23286 Calmodulin n=1 Tax=Candida albicans RepID=CALM_... 128 2e-28
UniRef100_Q295M8 GA14657 n=1 Tax=Drosophila pseudoobscura pseudo... 124 3e-28
UniRef100_B4GFY4 GL21535 n=1 Tax=Drosophila persimilis RepID=B4G... 124 3e-28
UniRef100_Q5VIR9 Calmodulin (Fragment) n=9 Tax=Penicillium RepID... 124 3e-28
UniRef100_B3FC25 Calmodulin (Fragment) n=1 Tax=Eurotium rubrum R... 125 3e-28
UniRef100_B3G4W6 Calmodulin (Fragment) n=1 Tax=Stylaster roseus ... 127 4e-28
UniRef100_B4M4P6 GJ10193 n=1 Tax=Drosophila virilis RepID=B4M4P6... 121 5e-28
UniRef100_UPI00015B4F26 PREDICTED: similar to calmodulin n=1 Tax... 122 6e-28
UniRef100_A9XEY8 Calmodulin (Fragment) n=63 Tax=Trichocomaceae R... 124 6e-28
UniRef100_Q32W24 Calmodulin (Fragment) n=1 Tax=Bonneviella sp. 3... 127 7e-28
UniRef100_Q32W17 Calmodulin (Fragment) n=1 Tax=Clytia gracilis R... 127 7e-28
UniRef100_B1NN71 Calmodulin (Fragment) n=22 Tax=Pezizomycotina R... 124 8e-28
UniRef100_C5XN41 Putative uncharacterized protein Sb03g037630 n=... 123 1e-27
UniRef100_Q9XZP3 Calmodulin-like protein n=1 Tax=Branchiostoma f... 120 1e-27
UniRef100_A9XEX0 Calmodulin (Fragment) n=1 Tax=Penicillium sp. N... 124 1e-27
UniRef100_B1NMV5 Calmodulin (Fragment) n=72 Tax=Trichocomaceae R... 125 1e-27
UniRef100_Q0IUU4 Putative calmodulin-like protein 2 n=1 Tax=Oryz... 121 2e-27
UniRef100_B4JW63 GH22800 n=1 Tax=Drosophila grimshawi RepID=B4JW... 125 2e-27
UniRef100_B3FHA2 Calmodulin (Fragment) n=1 Tax=Aspergillus ostia... 122 2e-27
UniRef100_O24033 Calmodulin (Fragment) n=1 Tax=Solanum lycopersi... 125 3e-27
UniRef100_B1NDP5 Calmodulin n=1 Tax=Actinidia deliciosa var. del... 120 3e-27
UniRef100_B0WSH2 Calmodulin n=1 Tax=Culex quinquefasciatus RepID... 116 4e-27
UniRef100_Q7QCA9 AGAP002536-PA (Fragment) n=1 Tax=Anopheles gamb... 116 4e-27
UniRef100_Q174F0 Calmodulin n=1 Tax=Aedes aegypti RepID=Q174F0_A... 116 4e-27
UniRef100_UPI000186E634 calmodulin-A, putative n=1 Tax=Pediculus... 120 4e-27
UniRef100_Q9NAS1 Calmodulin-like protein 2 n=1 Tax=Branchiostoma... 122 4e-27
UniRef100_A7T043 Predicted protein n=1 Tax=Nematostella vectensi... 117 4e-27
UniRef100_UPI000058840E PREDICTED: similar to calmodulin 2 n=1 T... 120 4e-27
UniRef100_B8BLX3 Putative uncharacterized protein n=1 Tax=Oryza ... 120 5e-27
UniRef100_UPI0000D9C039 PREDICTED: similar to calmodulin 1 n=1 T... 121 5e-27
UniRef100_B3FE91 Calmodulin (Fragment) n=1 Tax=Petromyces alliac... 121 5e-27
UniRef100_C1N698 Predicted protein n=1 Tax=Micromonas pusilla CC... 124 6e-27
UniRef100_C7BAI8 Calmodulin (Fragment) n=1 Tax=Calyptogena magni... 124 6e-27
UniRef100_Q0CZZ8 Calmodulin n=1 Tax=Aspergillus terreus NIH2624 ... 124 6e-27
UniRef100_UPI000186EA04 calmodulin, putative n=1 Tax=Pediculus h... 122 7e-27
UniRef100_C3XWH7 Putative uncharacterized protein n=1 Tax=Branch... 115 7e-27
UniRef100_B5G4N1 Putative calmodulin variant 1 n=1 Tax=Taeniopyg... 119 7e-27
UniRef100_C4R1N7 Calmodulin n=1 Tax=Pichia pastoris GS115 RepID=... 123 7e-27
UniRef100_B4ND50 GK10165 n=1 Tax=Drosophila willistoni RepID=B4N... 114 8e-27
UniRef100_B4MD26 GJ15350 n=1 Tax=Drosophila virilis RepID=B4MD26... 114 8e-27
UniRef100_Q29IW8 GA11114 n=1 Tax=Drosophila pseudoobscura pseudo... 114 8e-27
UniRef100_B3MYE7 GF22040 n=1 Tax=Drosophila ananassae RepID=B3MY... 114 8e-27
UniRef100_B4PXK0 GE16550 n=1 Tax=Drosophila yakuba RepID=B4PXK0_... 114 8e-27
UniRef100_B4K4U3 GI10339 n=1 Tax=Drosophila mojavensis RepID=B4K... 117 9e-27
UniRef100_C7FES6 Calmodulin (Fragment) n=1 Tax=Eurotium rubrum R... 117 9e-27
UniRef100_B3FDF6 Calmodulin (Fragment) n=1 Tax=Aspergillus tubin... 120 9e-27
UniRef100_B5THA2 Calmodulin 2 n=1 Tax=Euglena gracilis RepID=B5T... 123 1e-26
UniRef100_Q9NF73 EG:BACR7A4.12 protein n=1 Tax=Drosophila melano... 114 1e-26
UniRef100_B4I991 GM19002 n=1 Tax=Drosophila sechellia RepID=B4I9... 114 1e-26
UniRef100_Q8SYN0 CG11638 n=1 Tax=Drosophila melanogaster RepID=Q... 114 1e-26
UniRef100_Q96TN0 Calmodulin (Fragment) n=4 Tax=Pezizomycotina Re... 117 1e-26
UniRef100_C6ZJB5 UGT4 n=1 Tax=Pueraria montana var. lobata RepID... 122 1e-26
UniRef100_B1NNS4 Calmodulin (Fragment) n=3 Tax=Trichocomaceae Re... 122 1e-26
UniRef100_UPI0001792137 PREDICTED: similar to calmodulin n=1 Tax... 120 1e-26
UniRef100_Q3BDI8 Calmodulin-like protein n=1 Tax=Pinctada fucata... 118 1e-26
UniRef100_P04630 Calmodulin-like protein n=2 Tax=Caenorhabditis ... 114 1e-26
UniRef100_C1C2V8 Calmodulin n=1 Tax=Caligus clemensi RepID=C1C2V... 116 1e-26
UniRef100_B9RTI5 Calmodulin, putative n=1 Tax=Ricinus communis R... 117 1e-26
UniRef100_B9HKC0 Predicted protein n=1 Tax=Populus trichocarpa R... 117 1e-26
UniRef100_Q9ATG1 Calmodulin n=1 Tax=Castanea sativa RepID=Q9ATG1... 115 1e-26
UniRef100_Q0IQB6 Calmodulin-like protein 3 n=2 Tax=Oryza sativa ... 117 2e-26
UniRef100_Q39890 Putative uncharacterized protein n=1 Tax=Glycin... 116 2e-26
UniRef100_C6T231 Putative uncharacterized protein n=1 Tax=Glycin... 114 2e-26
UniRef100_B3FE94 Calmodulin (Fragment) n=32 Tax=Pezizomycotina R... 119 2e-26
UniRef100_UPI00003C0413 PREDICTED: similar to CG11638-PA isoform... 113 2e-26
UniRef100_C4JBJ3 Putative uncharacterized protein n=1 Tax=Zea ma... 120 2e-26
UniRef100_B9GBR1 Putative uncharacterized protein n=1 Tax=Oryza ... 117 2e-26
UniRef100_C6T1B7 Putative uncharacterized protein n=1 Tax=Glycin... 116 2e-26
UniRef100_UPI000186176E hypothetical protein BRAFLDRAFT_72667 n=... 121 3e-26
UniRef100_Q9NAS0 Calmodulin-like protein 3 (Fragment) n=1 Tax=Br... 121 3e-26
UniRef100_B4JKV0 GH12737 n=1 Tax=Drosophila grimshawi RepID=B4JK... 113 3e-26
UniRef100_C5YRF9 Putative uncharacterized protein Sb08g020710 n=... 114 3e-26
UniRef100_B7Q370 Nonmuscle myosin essential light chain, putativ... 117 3e-26
UniRef100_Q948R0 Calmodulin-like protein 5 n=2 Tax=Oryza sativa ... 114 4e-26
UniRef100_C6T2Y6 Putative uncharacterized protein n=1 Tax=Glycin... 116 4e-26
UniRef100_Q94IG4 Calmodulin NtCaM13 n=1 Tax=Nicotiana tabacum Re... 114 5e-26
UniRef100_Q43447 Calmodulin n=1 Tax=Glycine max RepID=Q43447_SOYBN 113 5e-26
UniRef100_A5BQ65 Chromosome chr5 scaffold_2, whole genome shotgu... 115 5e-26
UniRef100_C7BAI9 Calmodulin (Fragment) n=1 Tax=Vesicomya gigas R... 120 6e-26
UniRef100_B1NNT0 Calmodulin (Fragment) n=4 Tax=Pezizomycotina Re... 120 6e-26
UniRef100_B1NNL9 Calmodulin (Fragment) n=1 Tax=Aspergillus fumig... 120 6e-26
UniRef100_B1NNL8 Calmodulin (Fragment) n=3 Tax=Trichocomaceae Re... 120 6e-26
UniRef100_B1NNL7 Calmodulin (Fragment) n=1 Tax=Monascus purpureu... 120 6e-26
UniRef100_B1NNL5 Calmodulin (Fragment) n=27 Tax=Pezizomycotina R... 120 6e-26
UniRef100_UPI0000D56477 PREDICTED: similar to CG11638 CG11638-PA... 113 7e-26
UniRef100_B9HUQ2 Predicted protein n=1 Tax=Populus trichocarpa R... 114 7e-26
UniRef100_B9S4P7 Calmodulin, putative n=1 Tax=Ricinus communis R... 116 7e-26
UniRef100_B4PT12 GE23620 n=1 Tax=Drosophila yakuba RepID=B4PT12_... 115 7e-26
UniRef100_UPI0001863FA1 hypothetical protein BRAFLDRAFT_59201 n=... 120 8e-26
UniRef100_B7P3X2 Calmodulin, putative n=1 Tax=Ixodes scapularis ... 120 8e-26
UniRef100_P30187 Calmodulin-like protein 10 n=1 Tax=Arabidopsis ... 105 9e-26
UniRef100_UPI000155519F PREDICTED: hypothetical protein n=1 Tax=... 117 1e-25
UniRef100_C3ZXF1 Putative uncharacterized protein (Fragment) n=1... 117 1e-25
UniRef100_B6U6F8 Calmodulin-related protein n=1 Tax=Zea mays Rep... 119 1e-25
UniRef100_A8QF16 Calmodulin-like protein, putative n=1 Tax=Brugi... 111 2e-25
UniRef100_A7Q610 Chromosome chr14 scaffold_54, whole genome shot... 114 2e-25
UniRef100_B3P6E7 GG11425 n=1 Tax=Drosophila erecta RepID=B3P6E7_... 114 2e-25
UniRef100_Q17TS6 Calmodulin (Fragment) n=1 Tax=Penicillium waksm... 119 2e-25
UniRef100_B8MH94 Troponin C, putative n=1 Tax=Talaromyces stipit... 119 2e-25
UniRef100_Q9LIK5 Calmodulin-like protein 11 n=1 Tax=Arabidopsis ... 112 2e-25
UniRef100_B8QQE3 Calmodulin (Fragment) n=1 Tax=Eupenicillium sin... 118 2e-25
UniRef100_UPI00017E10DB hypothetical protein LOC100123509 n=1 Ta... 111 3e-25
UniRef100_UPI00017923EF PREDICTED: similar to AGAP002536-PA n=1 ... 111 3e-25
UniRef100_Q40982 Calmodulin-like protein n=1 Tax=Pisum sativum R... 112 3e-25
UniRef100_Q1W3B0 Calmodulin n=1 Tax=Striga asiatica RepID=Q1W3B0... 118 3e-25
UniRef100_C6T303 Putative uncharacterized protein n=1 Tax=Glycin... 113 3e-25
UniRef100_C1BLP2 Calmodulin n=1 Tax=Osmerus mordax RepID=C1BLP2_... 117 4e-25
UniRef100_C5Y416 Putative uncharacterized protein Sb05g002010 n=... 117 4e-25
UniRef100_C7BAJ0 Calmodulin (Fragment) n=1 Tax=Vesicomya pacific... 117 4e-25
UniRef100_Q17TT1 Calmodulin (Fragment) n=1 Tax=Penicillium steck... 117 4e-25
UniRef100_UPI000186ABFC hypothetical protein BRAFLDRAFT_110648 n... 114 4e-25
UniRef100_P06787 Calmodulin n=4 Tax=Saccharomyces cerevisiae Rep... 117 5e-25
UniRef100_A7SRU7 Predicted protein n=1 Tax=Nematostella vectensi... 112 6e-25
UniRef100_A3FBF5 Calmodulin n=1 Tax=Porphyra yezoensis RepID=A3F... 115 6e-25
UniRef100_B4QVF0 GD21235 n=2 Tax=melanogaster subgroup RepID=B4Q... 112 6e-25
UniRef100_P49258 Calmodulin-related protein 97A n=1 Tax=Drosophi... 112 8e-25
UniRef100_B3FC33 Calmodulin (Fragment) n=1 Tax=Eurotium amstelod... 116 9e-25
UniRef100_Q18092 Protein C18E9.1, confirmed by transcript eviden... 110 1e-24
UniRef100_P90620 Calmodulin (Fragment) n=1 Tax=Trichomonas vagin... 113 1e-24
UniRef100_O24034 Calmodulin (Fragment) n=1 Tax=Solanum lycopersi... 116 1e-24
UniRef100_C7G360 Calmodulin (Fragment) n=4 Tax=mitosporic Tricho... 116 1e-24
UniRef100_C5DUF6 ZYRO0C16346p n=1 Tax=Zygosaccharomyces rouxii C... 116 1e-24
UniRef100_O23320 Calmodulin-like protein 8 n=1 Tax=Arabidopsis t... 110 1e-24
UniRef100_B2AKK9 Predicted CDS Pa_5_8030 n=1 Tax=Podospora anser... 110 1e-24
UniRef100_B4K4U4 GI10340 n=1 Tax=Drosophila mojavensis RepID=B4K... 107 1e-24
UniRef100_Q3SDW5 Calmodulin 6-1 n=1 Tax=Paramecium tetraurelia R... 113 1e-24
UniRef100_B6U186 Calmodulin n=1 Tax=Zea mays RepID=B6U186_MAIZE 115 2e-24
UniRef100_A7TRW4 Putative uncharacterized protein n=1 Tax=Vander... 115 2e-24
UniRef100_B6DQN2 Putative calmodulin (Fragment) n=1 Tax=Taeniopy... 115 2e-24
UniRef100_Q75BA4 ADL337Wp n=1 Tax=Eremothecium gossypii RepID=Q7... 115 2e-24
UniRef100_Q6FWB6 Similar to uniprot|P06787 Saccharomyces cerevis... 115 2e-24
UniRef100_C5DL75 KLTH0F10626p n=1 Tax=Lachancea thermotolerans C... 115 2e-24
UniRef100_B6HRG1 Pc22g12180 protein n=1 Tax=Penicillium chrysoge... 115 2e-24
UniRef100_A8BNY0 Calmodulin n=3 Tax=Giardia intestinalis RepID=A... 114 2e-24
UniRef100_A8WSI9 C. briggsae CBR-CAL-2 protein n=1 Tax=Caenorhab... 109 3e-24
UniRef100_Q178H5 Calmodulin n=1 Tax=Aedes aegypti RepID=Q178H5_A... 106 3e-24
UniRef100_A8C1R9 Calmodulin (Fragment) n=8 Tax=Davidiellaceae Re... 114 3e-24
UniRef100_A8C1N7 Calmodulin (Fragment) n=1 Tax=Cladosporium aff.... 114 3e-24
UniRef100_B5E054 GA24239 n=1 Tax=Drosophila pseudoobscura pseudo... 114 4e-24