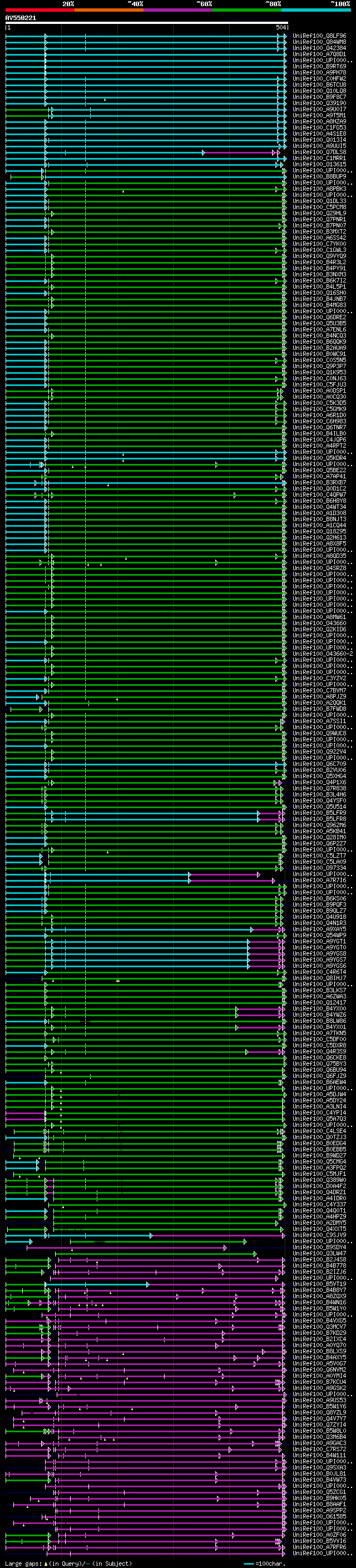
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV550221 RZ109h06R
(504 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q8LF96 PRL1 protein n=1 Tax=Arabidopsis thaliana RepID... 304 3e-90
UniRef100_Q84WM8 Putative uncharacterized protein At4g15900 n=1 ... 304 3e-90
UniRef100_Q42384 PP1/PP2A phosphatases pleiotropic regulator PRL... 304 3e-90
UniRef100_A7Q8D1 Chromosome chr5 scaffold_64, whole genome shotg... 288 9e-84
UniRef100_UPI00019831CE PREDICTED: hypothetical protein n=1 Tax=... 288 9e-84
UniRef100_B9RT69 PP1/PP2A phosphatases pleiotropic regulator PRL... 284 6e-83
UniRef100_A9PH78 Predicted protein n=1 Tax=Populus trichocarpa R... 282 5e-82
UniRef100_C0HFW2 Putative uncharacterized protein n=1 Tax=Zea ma... 273 2e-79
UniRef100_B6TCU8 PP1/PP2A phosphatases pleiotropic regulator PRL... 273 2e-79
UniRef100_Q10LQ8 Os03g0339100 protein n=2 Tax=Oryza sativa Japon... 270 1e-78
UniRef100_B9F8C7 Putative uncharacterized protein n=2 Tax=Oryza ... 264 6e-77
UniRef100_Q39190 PP1/PP2A phosphatases pleiotropic regulator PRL... 260 2e-76
UniRef100_A9U0I7 Predicted protein n=1 Tax=Physcomitrella patens... 257 2e-74
UniRef100_A9T5M1 Predicted protein (Fragment) n=1 Tax=Physcomitr... 257 2e-74
UniRef100_A8HZA9 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 224 3e-64
UniRef100_C1FG53 Predicted protein n=1 Tax=Micromonas sp. RCC299... 221 4e-63
UniRef100_A4S1E8 Predicted protein n=1 Tax=Ostreococcus lucimari... 221 1e-62
UniRef100_Q013I4 PRL1 (ISS) n=1 Tax=Ostreococcus tauri RepID=Q01... 221 2e-62
UniRef100_A9UUI5 Predicted protein n=1 Tax=Monosiga brevicollis ... 216 3e-60
UniRef100_Q7DLS8 PRL1 protein (Fragment) n=1 Tax=Arabidopsis tha... 203 5e-60
UniRef100_C1MRR1 Predicted protein (Fragment) n=1 Tax=Micromonas... 209 3e-59
UniRef100_O13615 Pre-mRNA-splicing factor prp46 n=1 Tax=Schizosa... 205 2e-58
UniRef100_UPI00015B514E PREDICTED: similar to striatin, putative... 206 2e-57
UniRef100_B8BUP9 Putative uncharacterized protein (Fragment) n=1... 214 7e-57
UniRef100_UPI00017929EC PREDICTED: similar to pleiotropic regula... 205 9e-57
UniRef100_A8PBK3 Putative uncharacterized protein n=1 Tax=Coprin... 203 9e-57
UniRef100_UPI0000D5571C PREDICTED: similar to Transport and Golg... 204 1e-56
UniRef100_Q1DL33 Putative uncharacterized protein n=1 Tax=Coccid... 199 1e-56
UniRef100_C5PCM8 Pre-mRNA splicing protein prp5, putative n=1 Ta... 199 1e-56
UniRef100_Q29HL9 GA14743 n=2 Tax=pseudoobscura subgroup RepID=Q2... 201 2e-56
UniRef100_Q7PNR1 AGAP005631-PA n=1 Tax=Anopheles gambiae RepID=Q... 204 2e-56
UniRef100_B7PN07 Guanine nucleotide-binding protein, putative (F... 202 3e-56
UniRef100_B3MXT2 GF19378 n=1 Tax=Drosophila ananassae RepID=B3MX... 201 3e-56
UniRef100_A6SS42 Putative uncharacterized protein n=1 Tax=Botryo... 198 3e-56
UniRef100_C7YK00 Predicted protein n=1 Tax=Nectria haematococca ... 199 3e-56
UniRef100_C1GWL3 Pre-mRNA-splicing factor prp46 n=1 Tax=Paracocc... 199 3e-56
UniRef100_Q9VYQ9 Transport and golgi organization 4 n=1 Tax=Dros... 201 4e-56
UniRef100_B4R3L2 GD17060 n=1 Tax=Drosophila simulans RepID=B4R3L... 201 4e-56
UniRef100_B4PY91 GE16491 n=1 Tax=Drosophila yakuba RepID=B4PY91_... 202 4e-56
UniRef100_B3NXM3 GG17704 n=1 Tax=Drosophila erecta RepID=B3NXM3_... 202 4e-56
UniRef100_B6K7I2 Pre-mRNA-splicing factor prp46 n=1 Tax=Schizosa... 201 4e-56
UniRef100_B4L5P1 GI21768 n=1 Tax=Drosophila mojavensis RepID=B4L... 201 6e-56
UniRef100_Q16SH0 Striatin, putative (Fragment) n=1 Tax=Aedes aeg... 202 6e-56
UniRef100_B4JNB7 GH24167 n=1 Tax=Drosophila grimshawi RepID=B4JN... 201 6e-56
UniRef100_B4MG83 GJ18540 n=1 Tax=Drosophila virilis RepID=B4MG83... 201 6e-56
UniRef100_UPI000023E0E7 hypothetical protein FG01337.1 n=1 Tax=G... 199 6e-56
UniRef100_Q6DRE2 Pleiotropic regulator 1 n=1 Tax=Danio rerio Rep... 201 7e-56
UniRef100_Q5U3B5 Plrg1 protein n=1 Tax=Danio rerio RepID=Q5U3B5_... 201 7e-56
UniRef100_A7ENL6 Putative uncharacterized protein n=1 Tax=Sclero... 197 7e-56
UniRef100_B4NCQ3 GK10085 n=1 Tax=Drosophila willistoni RepID=B4N... 201 7e-56
UniRef100_B6QQK9 mRNA splicing protein (Prp5), putative n=1 Tax=... 200 7e-56
UniRef100_B2AUA9 Predicted CDS Pa_1_18510 n=1 Tax=Podospora anse... 198 1e-55
UniRef100_B0WC91 Pleiotropic regulator 1 n=1 Tax=Culex quinquefa... 201 1e-55
UniRef100_C0S5N5 Pre-mRNA-splicing factor prp46 n=2 Tax=Paracocc... 199 1e-55
UniRef100_Q9P3P7 Probable pleiotropic regulator 1 (PLRG1) n=1 Ta... 197 1e-55
UniRef100_Q1K953 Pre-mRNA splicing factor prp46 n=1 Tax=Neurospo... 197 1e-55
UniRef100_C0NJ63 Pre-mRNA-splicing factor n=1 Tax=Ajellomyces ca... 197 1e-55
UniRef100_C5FJU3 Pre-mRNA splicing factor prp46 n=1 Tax=Microspo... 196 1e-55
UniRef100_A0DSP1 Chromosome undetermined scaffold_62, whole geno... 203 2e-55
UniRef100_A0CQ30 Chromosome undetermined scaffold_24, whole geno... 203 2e-55
UniRef100_C5K3D5 Pre-mRNA splicing factor prp46 n=1 Tax=Ajellomy... 197 2e-55
UniRef100_C5GMK9 Pre-mRNA splicing factor prp46 n=1 Tax=Ajellomy... 197 2e-55
UniRef100_A6R1D0 Pre-mRNA splicing factor prp46 n=1 Tax=Ajellomy... 197 2e-55
UniRef100_C6H983 Pre-mRNA-splicing factor prp46 n=1 Tax=Ajellomy... 196 3e-55
UniRef100_Q6TNR7 Pleiotropic regulator 1 n=1 Tax=Danio rerio Rep... 199 3e-55
UniRef100_B4ILB0 GM13251 n=1 Tax=Drosophila sechellia RepID=B4IL... 199 3e-55
UniRef100_C4JQP6 Pre-mRNA splicing factor prp46 n=1 Tax=Uncinoca... 196 3e-55
UniRef100_A4RPT2 Putative uncharacterized protein n=1 Tax=Magnap... 197 5e-55
UniRef100_UPI000042CB68 hypothetical protein CNBG1700 n=1 Tax=Cr... 197 5e-55
UniRef100_Q5KDR4 Pre-mRNA-splicing factor PRP46 n=1 Tax=Filobasi... 197 5e-55
UniRef100_UPI000051A5AB PREDICTED: similar to CG1796-PA n=1 Tax=... 200 5e-55
UniRef100_Q5BE22 Pre-mRNA-splicing factor prp46 n=2 Tax=Emericel... 196 6e-55
UniRef100_A7AP41 WD domain, G-beta repeat containing protein n=1... 203 8e-55
UniRef100_B3RXB7 Putative uncharacterized protein (Fragment) n=1... 197 8e-55
UniRef100_Q0D1C2 Pre-mRNA splicing factor prp46 n=1 Tax=Aspergil... 194 8e-55
UniRef100_C4QPW7 WD-repeat protein, putative n=1 Tax=Schistosoma... 197 8e-55
UniRef100_B6H8Y8 Pc16g04580 protein n=1 Tax=Penicillium chrysoge... 195 1e-54
UniRef100_Q4WT34 Pre-mRNA-splicing factor prp46 n=2 Tax=Aspergil... 195 1e-54
UniRef100_A1D308 MRNA splicing protein (Prp5), putative n=1 Tax=... 194 2e-54
UniRef100_B8NJT3 mRNA splicing protein (Prp5), putative n=2 Tax=... 194 2e-54
UniRef100_A1CQ44 mRNA splicing protein (Prp5), putative n=1 Tax=... 194 2e-54
UniRef100_Q18295 Protein D1054.15, confirmed by transcript evide... 199 3e-54
UniRef100_Q2H613 Putative uncharacterized protein n=1 Tax=Chaeto... 194 5e-54
UniRef100_A8X8F5 C. briggsae CBR-TAG-135 protein n=1 Tax=Caenorh... 199 7e-54
UniRef100_UPI00001222B8 Hypothetical protein CBG09714 n=1 Tax=Ca... 199 7e-54
UniRef100_A8QD35 Putative uncharacterized protein n=1 Tax=Malass... 192 9e-54
UniRef100_UPI000186E9F2 Pleiotropic regulator, putative n=1 Tax=... 196 9e-54
UniRef100_Q4SRZ8 Chromosome 18 SCAF14485, whole genome shotgun s... 193 1e-53
UniRef100_UPI0000D9B2A4 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_UPI00005A2F74 PREDICTED: similar to Pleiotropic regula... 194 2e-53
UniRef100_UPI0000610C92 PREDICTED: hypothetical protein n=1 Tax=... 194 2e-53
UniRef100_UPI000155DD4C PREDICTED: similar to Pleiotropic regula... 194 2e-53
UniRef100_UPI0000EB26D3 Pleiotropic regulator 1. n=1 Tax=Canis l... 194 2e-53
UniRef100_UPI00004E80C2 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_UPI00016E7841 UPI00016E7841 related cluster n=1 Tax=Ta... 194 2e-53
UniRef100_A8MW61 Putative uncharacterized protein PLRG1 n=1 Tax=... 194 2e-53
UniRef100_O43660 Pleiotropic regulator 1 n=1 Tax=Homo sapiens Re... 194 2e-53
UniRef100_Q2KID6 Pleiotropic regulator 1 n=1 Tax=Bos taurus RepI... 194 2e-53
UniRef100_UPI0000E20605 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_UPI00016E7840 UPI00016E7840 related cluster n=1 Tax=Ta... 194 2e-53
UniRef100_UPI0000E20604 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_O43660-2 Isoform 2 of Pleiotropic regulator 1 n=1 Tax=... 194 2e-53
UniRef100_UPI00018677A3 hypothetical protein BRAFLDRAFT_283267 n... 195 2e-53
UniRef100_UPI0000E20603 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_UPI0000E20607 PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_C3YZV2 Putative uncharacterized protein n=1 Tax=Branch... 195 2e-53
UniRef100_UPI000194C2BE PREDICTED: pleiotropic regulator 1 (PRL1... 194 2e-53
UniRef100_C7BVM7 Putative Pre-mRNA splicing protein n=1 Tax=Angi... 196 3e-53
UniRef100_A8PJZ9 Pre-mRNA splicing protein prp5, putative n=1 Ta... 200 3e-53
UniRef100_A2QQK1 Contig An08c0100, complete genome n=1 Tax=Asper... 190 3e-53
UniRef100_B7FWD8 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 203 4e-53
UniRef100_UPI0000F2D68E PREDICTED: hypothetical protein n=1 Tax=... 192 6e-53
UniRef100_A7SSI1 Predicted protein n=1 Tax=Nematostella vectensi... 193 6e-53
UniRef100_UPI000155B996 PREDICTED: hypothetical protein n=1 Tax=... 198 6e-53
UniRef100_Q9WUC8 Pleiotropic regulator 1 n=1 Tax=Rattus norvegic... 192 6e-53
UniRef100_UPI0001923854 PREDICTED: similar to predicted protein ... 192 6e-53
UniRef100_UPI000155D0AF PREDICTED: similar to LOC495399 protein ... 193 7e-53
UniRef100_Q922V4 Pleiotropic regulator 1 n=1 Tax=Mus musculus Re... 192 7e-53
UniRef100_UPI000164C44C UPI000164C44C related cluster n=1 Tax=Mu... 192 7e-53
UniRef100_Q6C709 Pre-mRNA-splicing factor PRP46 n=1 Tax=Yarrowia... 192 9e-53
UniRef100_B2VU06 Pre-mRNA-splicing factor prp46 n=1 Tax=Pyrenoph... 191 1e-52
UniRef100_Q5XHG4 LOC495006 protein n=1 Tax=Xenopus laevis RepID=... 192 2e-52
UniRef100_Q4P1X6 Putative uncharacterized protein n=1 Tax=Ustila... 188 3e-52
UniRef100_Q7R838 Plasmodium vivax PV1H14040_P n=1 Tax=Plasmodium... 194 3e-52
UniRef100_B3L4H6 Regulatory protein, putative n=1 Tax=Plasmodium... 194 3e-52
UniRef100_Q4YSF0 Regulatory protein, putative (Fragment) n=1 Tax... 194 3e-52
UniRef100_Q5U514 LOC495399 protein n=1 Tax=Xenopus laevis RepID=... 191 3e-52
UniRef100_B5LFR9 Tango4 (Fragment) n=1 Tax=Drosophila simulans R... 189 3e-52
UniRef100_B5LFR8 Tango4 (Fragment) n=1 Tax=Drosophila simulans R... 189 3e-52
UniRef100_Q962M6 PV1H14040_P n=1 Tax=Plasmodium vivax RepID=Q962... 194 4e-52
UniRef100_A5KB41 Splicing regulatory protein, putative n=1 Tax=P... 194 4e-52
UniRef100_Q28IM0 Pleiotropic regulator 1 (PRL1 homolog, Arabidop... 191 4e-52
UniRef100_Q6P2Z7 LOC394977 protein (Fragment) n=1 Tax=Xenopus (S... 191 4e-52
UniRef100_UPI000180CA5D PREDICTED: similar to pleiotropic regula... 186 5e-52
UniRef100_C5LZT7 Pleiotropic regulator, putative n=1 Tax=Perkins... 186 8e-52
UniRef100_C5LA09 Pleiotropic regulator, putative n=1 Tax=Perkins... 186 8e-52
UniRef100_O97334 Regulatory protein, putative n=1 Tax=Plasmodium... 194 1e-51
UniRef100_UPI0001985DD3 PREDICTED: hypothetical protein n=1 Tax=... 181 2e-51
UniRef100_A7R7I6 Chromosome undetermined scaffold_1811, whole ge... 181 2e-51
UniRef100_UPI0000E47B9C PREDICTED: similar to LOC394977 protein ... 185 2e-51
UniRef100_UPI0000E460E3 PREDICTED: similar to LOC394977 protein ... 185 2e-51
UniRef100_B6KS06 Pleiotropic regulator 1, putative n=1 Tax=Toxop... 189 8e-51
UniRef100_B9PQF3 Pleiotropic regulator, putative n=1 Tax=Toxopla... 189 8e-51
UniRef100_B9QLZ7 Pleiotropic regulator, putative n=1 Tax=Toxopla... 189 9e-51
UniRef100_Q4U918 Putative uncharacterized protein n=1 Tax=Theile... 191 2e-50
UniRef100_Q4N1R3 Putative uncharacterized protein n=1 Tax=Theile... 190 2e-50
UniRef100_A9XAY5 CG1796 (Fragment) n=1 Tax=Drosophila orena RepI... 182 4e-50
UniRef100_Q54WP9 WD40 repeat-containing protein n=1 Tax=Dictyost... 182 5e-50
UniRef100_A9YGT1 Transport and golgi organization 4 (Fragment) n... 181 7e-50
UniRef100_A9YGT0 Transport and golgi organization 4 (Fragment) n... 181 7e-50
UniRef100_A9YGS8 Transport and golgi organization 4 (Fragment) n... 181 7e-50
UniRef100_A9YGS7 Transport and golgi organization 4 (Fragment) n... 181 7e-50
UniRef100_A9YGS6 Transport and golgi organization 4 (Fragment) n... 181 7e-50
UniRef100_C4R6T4 Splicing factor that is found in the Cef1p subc... 182 9e-50
UniRef100_Q8IHJ7 Putative uncharacterized protein n=1 Tax=Brugia... 192 8e-48
UniRef100_UPI00006CAF04 hypothetical protein TTHERM_00684450 n=1... 174 7e-47
UniRef100_B3LKS7 Pre-mRNA splicing factor n=2 Tax=Saccharomyces ... 174 2e-46
UniRef100_A6ZWA3 Pre-mRNA splicing factor n=2 Tax=Saccharomyces ... 174 2e-46
UniRef100_Q12417 Pre-mRNA-splicing factor PRP46 n=1 Tax=Saccharo... 174 2e-46
UniRef100_B4YX00 Putative uncharacterized protein (Fragment) n=1... 168 3e-46
UniRef100_B4YWZ6 Putative uncharacterized protein (Fragment) n=1... 168 3e-46
UniRef100_B8LW86 mRNA splicing protein (Prp5), putative n=1 Tax=... 167 4e-46
UniRef100_B4YX01 Putative uncharacterized protein (Fragment) n=1... 167 8e-46
UniRef100_A7TKN5 Putative uncharacterized protein n=1 Tax=Vander... 172 5e-45
UniRef100_C5DF00 KLTH0D11132p n=1 Tax=Lachancea thermotolerans C... 167 8e-45
UniRef100_C5DXR8 ZYRO0F07282p n=1 Tax=Zygosaccharomyces rouxii C... 166 7e-44
UniRef100_Q4R3S9 Testis cDNA clone: QtsA-14439, similar to human... 162 9e-44
UniRef100_Q6CKE8 Pre-mRNA-splicing factor PRP46 n=1 Tax=Kluyvero... 166 1e-43
UniRef100_Q75BY3 Pre-mRNA-splicing factor PRP46 n=1 Tax=Eremothe... 165 2e-43
UniRef100_Q6BU94 Pre-mRNA-splicing factor PRP46 n=1 Tax=Debaryom... 162 2e-42
UniRef100_Q6FJZ9 Pre-mRNA-splicing factor PRP46 n=1 Tax=Candida ... 174 4e-42
UniRef100_B6AEW4 Putative uncharacterized protein n=1 Tax=Crypto... 160 5e-42
UniRef100_UPI00003BD789 hypothetical protein DEHA0C13607g n=1 Ta... 161 8e-42
UniRef100_A5DJW4 Putative uncharacterized protein n=1 Tax=Pichia... 155 7e-41
UniRef100_A5DY24 Pre-mRNA splicing factor PRP46 n=1 Tax=Lodderom... 161 7e-41
UniRef100_A3LNI4 Predicted protein n=1 Tax=Pichia stipitis RepID... 152 2e-40
UniRef100_C4YPI4 Pre-mRNA splicing factor PRP46 n=1 Tax=Candida ... 162 3e-40
UniRef100_Q5A7Q3 Pre-mRNA-splicing factor PRP46 n=1 Tax=Candida ... 162 3e-40
UniRef100_UPI000151BB74 hypothetical protein PGUG_03565 n=1 Tax=... 154 3e-40
UniRef100_C4LSE4 Pre-mRNA-splicing factor PRP46, putative n=1 Ta... 162 2e-39
UniRef100_Q0TZJ3 Putative uncharacterized protein n=1 Tax=Phaeos... 145 6e-39
UniRef100_B0EDG4 Pre-mRNA-splicing factor PRP46, putative n=1 Ta... 160 6e-39
UniRef100_B0EBB5 Mitogen-activated protein kinase organizer, put... 160 6e-39
UniRef100_B9WD27 Pre-mRNA-splicing factor, putative n=1 Tax=Cand... 162 9e-39
UniRef100_Q5CMG4 Pleiotropic regulator 1 n=1 Tax=Cryptosporidium... 145 8e-38
UniRef100_A3FPQ2 Pleiotropic regulator 1 n=1 Tax=Cryptosporidium... 147 1e-37
UniRef100_C5MJF1 Putative uncharacterized protein n=1 Tax=Candid... 157 4e-37
UniRef100_Q389W0 Putative uncharacterized protein n=1 Tax=Trypan... 142 4e-37
UniRef100_D0A4F2 Putative uncharacterized protein n=1 Tax=Trypan... 142 4e-37
UniRef100_Q4DRZ1 Putative uncharacterized protein n=1 Tax=Trypan... 142 9e-37
UniRef100_A4IDR0 Putative uncharacterized protein n=1 Tax=Leishm... 138 8e-36
UniRef100_C4Y337 Putative uncharacterized protein n=1 Tax=Clavis... 151 2e-35
UniRef100_Q4Q0T1 Putative uncharacterized protein n=1 Tax=Leishm... 136 4e-35
UniRef100_A4HPZ9 Putative uncharacterized protein n=1 Tax=Leishm... 134 4e-34
UniRef100_A2DMY5 Pre-mRNA splicing protein, putative n=1 Tax=Tri... 144 4e-33
UniRef100_Q4XXT5 Regulatory protein, putative (Fragment) n=1 Tax... 140 6e-33
UniRef100_C9SJV9 Pre-mRNA-splicing factor prp46 n=1 Tax=Verticil... 114 1e-30
UniRef100_UPI000187E9D8 hypothetical protein MPER_07995 n=1 Tax=... 122 2e-29
UniRef100_B9SDY4 Putative uncharacterized protein n=1 Tax=Ricinu... 119 1e-25
UniRef100_Q3LW47 mRNA splicing factor PRL1 n=1 Tax=Bigelowiella ... 117 4e-25
UniRef100_B2J4S8 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 109 1e-24
UniRef100_B4B778 Serine/threonine protein kinase with WD40 repea... 98 1e-23
UniRef100_B2IZJ6 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 100 5e-23
UniRef100_UPI000192562D PREDICTED: similar to AGAP005631-PA n=1 ... 110 7e-23
UniRef100_B5VT19 YPL151Cp-like protein (Fragment) n=1 Tax=Saccha... 94 2e-22
UniRef100_B4B8Y7 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 101 3e-21
UniRef100_A8ZQX9 WD repeat protein n=1 Tax=Acaryochloris marina ... 92 6e-21
UniRef100_B4WN16 Putative uncharacterized protein n=1 Tax=Synech... 99 6e-21
UniRef100_B5W1Y0 WD-40 repeat protein n=1 Tax=Arthrospira maxima... 91 1e-20
UniRef100_UPI000175830C PREDICTED: similar to katanin p80 subuni... 102 1e-20
UniRef100_B4VXG5 Putative uncharacterized protein n=1 Tax=Microc... 102 2e-20
UniRef100_Q3MCV7 Ribosome assembly protein 4 (RSA4) n=1 Tax=Anab... 101 2e-20
UniRef100_B7KD29 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 94 2e-20
UniRef100_B2IXC4 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 99 2e-20
UniRef100_A0YQ70 Serine/Threonine protein kinase with WD40 repea... 99 2e-20
UniRef100_B8LXS9 G-protein beta WD-40 repeats containing protein... 98 3e-20
UniRef100_B4AXY5 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 97 3e-20
UniRef100_A5V0G7 NB-ARC domain protein n=1 Tax=Roseiflexus sp. R... 97 4e-20
UniRef100_Q6NVM2 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 100 4e-20
UniRef100_A0YMI4 WD-40 repeat protein n=1 Tax=Lyngbya sp. PCC 81... 94 5e-20
UniRef100_B7KCU4 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 94 5e-20
UniRef100_A9GSK2 WD-repeat protein n=1 Tax=Sorangium cellulosum ... 92 6e-20
UniRef100_UPI00015B4285 PREDICTED: similar to ENSANGP00000021697... 100 7e-20
UniRef100_A9US53 Predicted protein n=1 Tax=Monosiga brevicollis ... 100 7e-20
UniRef100_B5W1Y6 WD-40 repeat protein n=1 Tax=Arthrospira maxima... 91 1e-19
UniRef100_Q8YZL9 Serine/threonine kinase with WD-40 repeat n=1 T... 99 1e-19
UniRef100_Q4V7Y7 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 99 1e-19
UniRef100_Q7ZYI4 Katnb1 protein n=1 Tax=Xenopus laevis RepID=Q7Z... 99 2e-19
UniRef100_B5W8L0 WD-40 repeat protein n=1 Tax=Arthrospira maxima... 94 2e-19
UniRef100_Q3M6B4 Serine/threonine protein kinase with WD40 repea... 99 2e-19
UniRef100_A9GAC3 Hypothetical WD-repeat protein n=1 Tax=Sorangiu... 99 2e-19
UniRef100_C7RS72 WD-40 repeat protein n=1 Tax=Candidatus Accumul... 99 2e-19
UniRef100_B4W111 Putative uncharacterized protein n=1 Tax=Microc... 99 2e-19
UniRef100_UPI0001738F97 nucleotide binding n=1 Tax=Arabidopsis t... 98 3e-19
UniRef100_Q9SXA3 T28P6.17 protein n=1 Tax=Arabidopsis thaliana R... 98 3e-19
UniRef100_B0JL81 Serine/threonine protein kinase n=1 Tax=Microcy... 94 3e-19
UniRef100_B4VW73 Protein kinase domain n=1 Tax=Microcoleus chtho... 96 3e-19
UniRef100_UPI0001927375 PREDICTED: similar to predicted protein ... 98 3e-19
UniRef100_Q5ZCG1 Os01g0780400 protein n=1 Tax=Oryza sativa Japon... 98 3e-19
UniRef100_B9HK05 Predicted protein n=1 Tax=Populus trichocarpa R... 98 3e-19
UniRef100_B8AAF1 Putative uncharacterized protein n=1 Tax=Oryza ... 98 3e-19
UniRef100_A9SPP2 Predicted protein (Fragment) n=1 Tax=Physcomitr... 98 3e-19
UniRef100_O61585 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 98 3e-19
UniRef100_UPI0001795DC3 PREDICTED: similar to Katanin p80 WD40-c... 97 5e-19
UniRef100_UPI000161F414 predicted protein n=1 Tax=Physcomitrella... 97 5e-19
UniRef100_A0ZF06 Serine/Threonine protein kinase with WD40 repea... 89 5e-19
UniRef100_B5VVI6 WD-40 repeat protein n=1 Tax=Arthrospira maxima... 91 5e-19
UniRef100_A7RFR6 Predicted protein (Fragment) n=1 Tax=Nematostel... 89 5e-19
UniRef100_UPI00016E107C UPI00016E107C related cluster n=1 Tax=Ta... 97 6e-19
UniRef100_UPI00016E107B UPI00016E107B related cluster n=1 Tax=Ta... 97 6e-19
UniRef100_B2J3M1 Heat shock protein DnaJ domain protein n=1 Tax=... 97 6e-19
UniRef100_B0C2Q9 WD-repeat protein n=1 Tax=Acaryochloris marina ... 97 6e-19
UniRef100_A8ZQA5 WD-repeat protein n=1 Tax=Acaryochloris marina ... 89 7e-19
UniRef100_Q3M972 Serine/threonine protein kinase with WD40 repea... 95 7e-19
UniRef100_B4B8Y6 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 94 7e-19
UniRef100_C5XLP5 Putative uncharacterized protein Sb03g036400 n=... 97 8e-19
UniRef100_B8LTB2 Pfs, NACHT and WD domain protein n=1 Tax=Talaro... 92 9e-19
UniRef100_UPI000051A267 PREDICTED: similar to katanin p80 (WD40-... 96 1e-18
UniRef100_A0CBM5 Chromosome undetermined scaffold_165, whole gen... 93 1e-18
UniRef100_B4VHM7 Putative uncharacterized protein n=1 Tax=Microc... 91 1e-18
UniRef100_A0ZIJ5 Putative uncharacterized protein n=1 Tax=Nodula... 92 1e-18
UniRef100_B1N3D9 Pre-mRNA-splicing factor PRP46, putative n=1 Ta... 92 1e-18
UniRef100_UPI000034F4C7 WD-40 repeat family protein / katanin p8... 96 1e-18
UniRef100_UPI00017B35BC UPI00017B35BC related cluster n=1 Tax=Te... 96 1e-18
UniRef100_O22725 F11P17.7 protein n=1 Tax=Arabidopsis thaliana R... 96 1e-18
UniRef100_B7K750 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 96 1e-18
UniRef100_A0ZIJ6 Serine/Threonine protein kinase with WD40 repea... 94 1e-18
UniRef100_UPI000176152B PREDICTED: similar to katanin p80 (WD re... 96 2e-18
UniRef100_UPI0000248F7F PREDICTED: similar to katanin p80 (WD re... 96 2e-18
UniRef100_B9HUZ5 Predicted protein n=1 Tax=Populus trichocarpa R... 96 2e-18
UniRef100_Q7ZUV2 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 96 2e-18
UniRef100_A7SN68 Predicted protein n=1 Tax=Nematostella vectensi... 92 2e-18
UniRef100_UPI0001985918 PREDICTED: hypothetical protein n=1 Tax=... 95 2e-18
UniRef100_UPI00018684C0 hypothetical protein BRAFLDRAFT_130320 n... 95 2e-18
UniRef100_A3IK50 Peptidase C14, caspase catalytic subunit p20 n=... 95 2e-18
UniRef100_A7QNQ6 Chromosome undetermined scaffold_133, whole gen... 95 2e-18
UniRef100_A5AVC7 Putative uncharacterized protein n=1 Tax=Vitis ... 95 2e-18
UniRef100_C3YLC5 Putative uncharacterized protein n=1 Tax=Branch... 95 2e-18
UniRef100_A0E6P3 Chromosome undetermined scaffold_8, whole genom... 90 2e-18
UniRef100_A7EJN8 Putative uncharacterized protein n=1 Tax=Sclero... 92 2e-18
UniRef100_B1WUP9 WD-repeat protein n=1 Tax=Cyanothece sp. ATCC 5... 89 2e-18
UniRef100_UPI00019860C3 PREDICTED: hypothetical protein n=1 Tax=... 95 3e-18
UniRef100_A7R3U5 Chromosome undetermined scaffold_569, whole gen... 95 3e-18
UniRef100_B4W0K8 Putative uncharacterized protein n=1 Tax=Microc... 88 3e-18
UniRef100_A7ERN6 Putative uncharacterized protein n=1 Tax=Sclero... 94 4e-18
UniRef100_UPI0000E242AF PREDICTED: katanin p80 subunit B 1 isofo... 94 5e-18
UniRef100_UPI0000D9F18B PREDICTED: katanin p80 subunit B 1 n=1 T... 94 5e-18
UniRef100_UPI00005A0223 PREDICTED: similar to Katanin p80 WD40-c... 94 5e-18
UniRef100_Q4VFZ4 Katanin p80 subunit B1 n=1 Tax=Rattus norvegicu... 94 5e-18
UniRef100_B1WBN6 Katnb1 protein (Fragment) n=1 Tax=Rattus norveg... 94 5e-18
UniRef100_Q8YSC0 All3169 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 94 5e-18
UniRef100_B2IUK9 Protein kinase n=1 Tax=Nostoc punctiforme PCC 7... 94 5e-18
UniRef100_B9S5Z3 Katanin P80 subunit, putative n=1 Tax=Ricinus c... 94 5e-18
UniRef100_A6QQU1 KATNB1 protein n=1 Tax=Bos taurus RepID=A6QQU1_... 94 5e-18
UniRef100_Q8BG40 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 94 5e-18
UniRef100_Q9BVA0 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 94 5e-18
UniRef100_A7EAT8 Putative uncharacterized protein n=1 Tax=Sclero... 90 5e-18
UniRef100_A0YWB3 Serine/Threonine protein kinase with WD40 repea... 93 5e-18
UniRef100_UPI000194CF8B PREDICTED: katanin p80 subunit B 1 n=1 T... 94 7e-18
UniRef100_A0CJ90 Chromosome undetermined scaffold_2, whole genom... 90 7e-18
UniRef100_B8MFB6 WD-repeat protein, putative n=1 Tax=Talaromyces... 82 7e-18
UniRef100_UPI000155D011 PREDICTED: similar to Katanin p80 (WD re... 93 9e-18
UniRef100_B0C7J8 WD-repeat protein n=1 Tax=Acaryochloris marina ... 93 9e-18
UniRef100_B5W8I9 FHA domain containing protein n=1 Tax=Arthrospi... 93 9e-18
UniRef100_Q3M407 WD-40 repeat n=1 Tax=Anabaena variabilis ATCC 2... 93 9e-18
UniRef100_B2J0L8 Protein kinase n=1 Tax=Nostoc punctiforme PCC 7... 93 1e-17
UniRef100_B0CFV1 WD-repeat protein n=1 Tax=Acaryochloris marina ... 93 1e-17
UniRef100_C1E4X3 Katanin p80 subunit-like protein n=1 Tax=Microm... 93 1e-17
UniRef100_A7SV12 Predicted protein n=1 Tax=Nematostella vectensi... 93 1e-17
UniRef100_Q8H0T9-2 Isoform 2 of Katanin p80 WD40 repeat-containi... 93 1e-17
UniRef100_Q8H0T9 Katanin p80 WD40 repeat-containing subunit B1 h... 93 1e-17
UniRef100_Q10XR9 WD-40 repeat n=1 Tax=Trichodesmium erythraeum I... 92 1e-17
UniRef100_B7K999 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 92 1e-17
UniRef100_A5URP9 Ribosome assembly protein 4 (RSA4) n=1 Tax=Rose... 89 1e-17
UniRef100_A0YQ89 Putative uncharacterized protein n=1 Tax=Lyngby... 88 1e-17
UniRef100_B4W4D8 DnaJ domain protein n=1 Tax=Microcoleus chthono... 90 1e-17
UniRef100_UPI0001505839 unknown protein n=1 Tax=Arabidopsis thal... 92 1e-17
UniRef100_Q9FT96 Katanin p80 subunit-like protein n=1 Tax=Arabid... 92 1e-17
UniRef100_Q0WV51 Katanin p80 subunit-like protein n=1 Tax=Arabid... 92 1e-17
UniRef100_A3IRL3 Peptidase C14, caspase catalytic subunit p20 n=... 84 2e-17
UniRef100_B0D541 Predicted protein n=1 Tax=Laccaria bicolor S238... 86 2e-17
UniRef100_A0YUL3 Peptidase C14, caspase catalytic subunit p20 n=... 90 2e-17
UniRef100_Q2H6X7 Putative uncharacterized protein n=1 Tax=Chaeto... 85 2e-17
UniRef100_A0EG21 Chromosome undetermined scaffold_94, whole geno... 87 2e-17
UniRef100_A0YT97 WD-40 repeat protein n=1 Tax=Lyngbya sp. PCC 81... 92 2e-17
UniRef100_UPI0000F2B9AF PREDICTED: similar to Katanin p80 (WD re... 92 2e-17
UniRef100_UPI0000DD9348 Os04g0677700 n=1 Tax=Oryza sativa Japoni... 92 2e-17
UniRef100_Q3M3I4 Serine/threonine protein kinase with WD40 repea... 92 2e-17
UniRef100_B1WPV3 WD-repeat protein n=1 Tax=Cyanothece sp. ATCC 5... 92 2e-17
UniRef100_Q7XKB3 OSJNBa0064G10.18 protein n=1 Tax=Oryza sativa J... 92 2e-17
UniRef100_Q259K2 H0402C08.11 protein n=1 Tax=Oryza sativa RepID=... 92 2e-17
UniRef100_Q259E8 H0801D08.1 protein n=1 Tax=Oryza sativa RepID=Q... 92 2e-17
UniRef100_B9FDB0 Putative uncharacterized protein n=1 Tax=Oryza ... 92 2e-17
UniRef100_B8ARG7 Putative uncharacterized protein n=1 Tax=Oryza ... 92 2e-17
UniRef100_B8HZ95 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 89 2e-17
UniRef100_B9YJ79 WD-40 repeat protein n=1 Tax='Nostoc azollae' 0... 90 2e-17
UniRef100_UPI0001869D5C hypothetical protein BRAFLDRAFT_131378 n... 91 2e-17
UniRef100_C3ZS55 Putative uncharacterized protein n=1 Tax=Branch... 91 2e-17
UniRef100_A8ZLP0 WD-40 repeat protein n=1 Tax=Acaryochloris mari... 80 2e-17
UniRef100_UPI000186CE82 Katanin p80 WD40-containing subunit B1, ... 92 2e-17
UniRef100_Q3M803 Pentapeptide repeat n=1 Tax=Anabaena variabilis... 92 2e-17
UniRef100_Q3M2E2 Serine/threonine protein kinase with WD40 repea... 92 2e-17
UniRef100_B8HUE8 Serine/threonine protein kinase with WD40 repea... 92 2e-17
UniRef100_Q8LNT7 Putative microtubule-severing protein subunit n... 92 2e-17
UniRef100_Q337H9 Os10g0494800 protein n=1 Tax=Oryza sativa Japon... 92 2e-17
UniRef100_B9RJ00 Katanin P80 subunit, putative n=1 Tax=Ricinus c... 92 2e-17
UniRef100_B9G6F5 Putative uncharacterized protein n=1 Tax=Oryza ... 92 2e-17
UniRef100_B8BHN3 Putative uncharacterized protein n=1 Tax=Oryza ... 92 2e-17
UniRef100_A7F664 Putative uncharacterized protein n=1 Tax=Sclero... 92 2e-17
UniRef100_A1BER4 Ribosome assembly protein 4 (RSA4) n=1 Tax=Chlo... 86 3e-17
UniRef100_C7QPN4 Pentapeptide repeat protein n=1 Tax=Cyanothece ... 83 3e-17
UniRef100_B8MPQ7 WD-repeat protein, putative n=1 Tax=Talaromyces... 86 3e-17
UniRef100_B4B5L8 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 90 3e-17
UniRef100_B5VYQ4 Serine/threonine protein kinase with WD40 repea... 91 3e-17
UniRef100_A0CLS6 Chromosome undetermined scaffold_205, whole gen... 86 3e-17
UniRef100_B0C4Z9 WD-repeat protein n=1 Tax=Acaryochloris marina ... 91 3e-17
UniRef100_B6BLP1 WD-40 repeat protein n=1 Tax=Campylobacterales ... 91 3e-17
UniRef100_A7IQW2 HNWD1 protein n=1 Tax=Podospora anserina RepID=... 85 3e-17
UniRef100_Q8YRI1 Uncharacterized WD repeat-containing protein al... 87 3e-17
UniRef100_B8GB84 NB-ARC domain protein n=1 Tax=Chloroflexus aggr... 85 3e-17
UniRef100_B8HM86 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 84 3e-17
UniRef100_A7EXQ2 Putative uncharacterized protein n=1 Tax=Sclero... 89 3e-17
UniRef100_A8YMC6 Similar to tr|Q8YSG6|Q8YSG6 n=1 Tax=Microcystis... 89 3e-17
UniRef100_A0YWA5 Putative uncharacterized protein n=1 Tax=Lyngby... 88 3e-17
UniRef100_Q2TX39 WD40 repeat n=1 Tax=Aspergillus oryzae RepID=Q2... 89 3e-17
UniRef100_A8ZLT0 WD-repeat protein n=1 Tax=Acaryochloris marina ... 91 4e-17
UniRef100_A7BTI4 G-protein beta WD-40 repeat n=1 Tax=Beggiatoa s... 91 4e-17
UniRef100_UPI0001792A0B PREDICTED: similar to wd-repeat protein ... 91 6e-17
UniRef100_Q2GV04 Putative uncharacterized protein n=1 Tax=Chaeto... 91 6e-17
UniRef100_B8MEW8 WD-repeat protein, putative n=1 Tax=Talaromyces... 91 6e-17
UniRef100_A0EB01 Chromosome undetermined scaffold_87, whole geno... 86 6e-17
UniRef100_B1WNU5 WD-40 repeat-containing serine/threonine protei... 87 6e-17
UniRef100_Q8YL09 WD-repeat protein n=1 Tax=Nostoc sp. PCC 7120 R... 90 7e-17
UniRef100_B4B4U5 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 90 7e-17
UniRef100_A0YXI8 WD-40 repeat protein n=1 Tax=Lyngbya sp. PCC 81... 90 7e-17
UniRef100_A0E7C7 Chromosome undetermined scaffold_81, whole geno... 90 7e-17
UniRef100_Q8YTC2 Uncharacterized WD repeat-containing protein al... 90 7e-17
UniRef100_B8LXS7 F-box and wd40 domain protein, putative n=1 Tax... 85 7e-17
UniRef100_A7EPC1 Putative uncharacterized protein n=1 Tax=Sclero... 87 8e-17
UniRef100_Q7NJ67 WD-repeat protein n=1 Tax=Gloeobacter violaceus... 90 9e-17
UniRef100_B0JPS2 WD-repeat protein n=1 Tax=Microcystis aeruginos... 90 9e-17
UniRef100_B4WTY0 Putative uncharacterized protein n=1 Tax=Synech... 90 9e-17
UniRef100_A0ZDS0 Serine/Threonine protein kinase with WD40 repea... 84 1e-16
UniRef100_B8MYS2 Wd40 protein, putative n=1 Tax=Aspergillus flav... 89 1e-16
UniRef100_UPI0001985F2C PREDICTED: hypothetical protein n=1 Tax=... 89 1e-16
UniRef100_A7Q5K1 Chromosome undetermined scaffold_53, whole geno... 89 1e-16
UniRef100_A8I2S8 Centriole proteome protein (Fragment) n=1 Tax=C... 84 1e-16
UniRef100_B5X168 U4/U6 small nuclear ribonucleoprotein Prp4 n=1 ... 89 2e-16
UniRef100_Q7ND80 WD-repeat protein n=1 Tax=Gloeobacter violaceus... 89 2e-16
UniRef100_A8ZKU8 WD-repeat protein n=1 Tax=Acaryochloris marina ... 89 2e-16
UniRef100_A9SQZ7 Predicted protein n=1 Tax=Physcomitrella patens... 89 2e-16
UniRef100_B2ALD4 Predicted CDS Pa_5_3370 n=1 Tax=Podospora anser... 89 2e-16
UniRef100_B2J557 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 88 2e-16
UniRef100_A0DHV3 Chromosome undetermined scaffold_502, whole gen... 84 2e-16
UniRef100_A0ZFT6 WD-40 repeat protein n=1 Tax=Nodularia spumigen... 82 2e-16
UniRef100_UPI000180BFEE PREDICTED: similar to katanin p80 subuni... 89 2e-16
UniRef100_UPI0000D9F8F8 PREDICTED: similar to pleiotropic regula... 89 2e-16
UniRef100_Q7ND85 WD-repeat protein n=1 Tax=Gloeobacter violaceus... 89 2e-16
UniRef100_B0JNY6 WD-repeat protein n=1 Tax=Microcystis aeruginos... 89 2e-16
UniRef100_B5VZL8 Serine/threonine protein kinase with WD40 repea... 89 2e-16
UniRef100_B4B707 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 89 2e-16
UniRef100_A0YUC6 Serine/threonine kinase with WD-40 repeat n=1 T... 89 2e-16
UniRef100_B9RGB3 WD-repeat protein, putative n=1 Tax=Ricinus com... 89 2e-16
UniRef100_B6QCX5 Pfs, NACHT and WD domain protein n=1 Tax=Penici... 85 2e-16
UniRef100_B2ADI5 Predicted CDS Pa_4_1190 n=1 Tax=Podospora anser... 85 2e-16
UniRef100_B8MIB7 G-protein beta WD-40 repeats containing protein... 82 2e-16
UniRef100_C7ZCX2 Putative uncharacterized protein n=1 Tax=Nectri... 84 2e-16
UniRef100_A0C273 Chromosome undetermined scaffold_144, whole gen... 87 2e-16
UniRef100_B3RIM3 Putative uncharacterized protein n=1 Tax=Tricho... 88 3e-16
UniRef100_A9V3J2 Predicted protein n=1 Tax=Monosiga brevicollis ... 88 3e-16
UniRef100_A2G3K8 WD repeat protein, putative n=1 Tax=Trichomonas... 88 3e-16
UniRef100_A0BEV4 Chromosome undetermined scaffold_102, whole gen... 88 3e-16
UniRef100_A7EU93 Putative uncharacterized protein n=1 Tax=Sclero... 88 3e-16
UniRef100_A1DJ78 Wd-repeat protein n=1 Tax=Neosartorya fischeri ... 81 3e-16
UniRef100_A0YUE4 WD-repeat protein n=1 Tax=Lyngbya sp. PCC 8106 ... 82 3e-16
UniRef100_B2J4D1 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 86 3e-16
UniRef100_A0E4X7 Chromosome undetermined scaffold_79, whole geno... 87 3e-16
UniRef100_C1BM69 WD repeat protein 69 n=1 Tax=Osmerus mordax Rep... 87 3e-16
UniRef100_A9FTS8 WD-repeat protein n=1 Tax=Sorangium cellulosum ... 88 4e-16
UniRef100_C1ZDX2 WD-40 repeat-containing protein n=1 Tax=Plancto... 88 4e-16
UniRef100_A7BLC5 WD-40 repeat protein n=1 Tax=Beggiatoa sp. SS R... 88 4e-16
UniRef100_A0ZIS9 WD-40 repeat protein n=1 Tax=Nodularia spumigen... 88 4e-16
UniRef100_B4LMS6 GJ20531 n=1 Tax=Drosophila virilis RepID=B4LMS6... 88 4e-16
UniRef100_A0CXK4 Chromosome undetermined scaffold_30, whole geno... 88 4e-16
UniRef100_Q2UR60 WD40 repeat n=1 Tax=Aspergillus oryzae RepID=Q2... 88 4e-16
UniRef100_B2ACC8 Predicted CDS Pa_3_630 (Fragment) n=1 Tax=Podos... 88 4e-16
UniRef100_A7IQV8 NWD2 protein n=1 Tax=Podospora anserina RepID=A... 88 4e-16
UniRef100_Q5ZIU8 Katanin p80 WD40-containing subunit B1 n=1 Tax=... 88 4e-16
UniRef100_B4AUL0 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 87 4e-16
UniRef100_B2AVF6 Predicted CDS Pa_7_3110 n=1 Tax=Podospora anser... 84 5e-16
UniRef100_UPI000194D47E PREDICTED: PRP4 pre-mRNA processing fact... 87 5e-16
UniRef100_UPI00016E5C64 UPI00016E5C64 related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C63 UPI00016E5C63 related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C62 UPI00016E5C62 related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C61 UPI00016E5C61 related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C60 UPI00016E5C60 related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C5F UPI00016E5C5F related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C5E UPI00016E5C5E related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C0B UPI00016E5C0B related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_UPI00016E5C0A UPI00016E5C0A related cluster n=1 Tax=Ta... 87 5e-16
UniRef100_B7K7B4 WD-40 repeat protein n=1 Tax=Cyanothece sp. PCC... 87 5e-16
UniRef100_A0YQZ5 WD-repeat protein n=1 Tax=Lyngbya sp. PCC 8106 ... 87 5e-16
UniRef100_B9GWW8 Predicted protein (Fragment) n=1 Tax=Populus tr... 87 5e-16
UniRef100_B8CEM0 Predicted protein (Fragment) n=1 Tax=Thalassios... 87 5e-16
UniRef100_B3RNA4 Putative uncharacterized protein n=1 Tax=Tricho... 87 5e-16
UniRef100_A7IQW0 HNWD3 protein n=1 Tax=Podospora anserina RepID=... 84 5e-16
UniRef100_A0CJ89 Chromosome undetermined scaffold_199, whole gen... 82 5e-16
UniRef100_Q2GT52 Putative uncharacterized protein n=1 Tax=Chaeto... 83 5e-16
UniRef100_B0JW93 Serine/threonine protein kinase with WD40 repea... 79 5e-16
UniRef100_B9H9H5 Predicted protein n=1 Tax=Populus trichocarpa R... 84 5e-16
UniRef100_A0D039 Chromosome undetermined scaffold_33, whole geno... 87 6e-16
UniRef100_C8VEI0 NACHT and WD40 domain protein (AFU_orthologue; ... 85 6e-16
UniRef100_B8MUB1 WD-repeat protein, putative n=1 Tax=Talaromyces... 78 6e-16
UniRef100_Q7NCT8 WD-repeat protein n=1 Tax=Gloeobacter violaceus... 83 6e-16
UniRef100_Q6PH45 PRP4 pre-mRNA processing factor 4 homolog (Yeas... 87 6e-16
UniRef100_Q4TC09 Chromosome undetermined SCAF7064, whole genome ... 87 6e-16
UniRef100_Q8YSG6 Alr3119 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 87 6e-16
UniRef100_B4WRJ5 Putative uncharacterized protein n=1 Tax=Synech... 87 6e-16
UniRef100_Q9VVI0 CG6322 n=1 Tax=Drosophila melanogaster RepID=Q9... 87 6e-16
UniRef100_B4QNW2 GD12400 n=1 Tax=Drosophila simulans RepID=B4QNW... 87 6e-16
UniRef100_B4PJU2 GE22143 n=1 Tax=Drosophila yakuba RepID=B4PJU2_... 87 6e-16
UniRef100_B4HKT7 GM24327 n=1 Tax=Drosophila sechellia RepID=B4HK... 87 6e-16
UniRef100_B3NHU2 GG15805 n=1 Tax=Drosophila erecta RepID=B3NHU2_... 87 6e-16
UniRef100_B8MQQ4 WD-repeat protein, putative n=1 Tax=Talaromyces... 83 6e-16
UniRef100_B9YJK3 WD-40 repeat protein n=1 Tax='Nostoc azollae' 0... 83 6e-16
UniRef100_B1X336 WD-repeat protein n=1 Tax=Cyanothece sp. ATCC 5... 83 6e-16
UniRef100_B8NN35 Vegetative incompatibility WD repeat protein, p... 79 6e-16
UniRef100_A8YGJ6 Genome sequencing data, contig C309 n=1 Tax=Mic... 87 8e-16
UniRef100_UPI0000E811D6 PREDICTED: hypothetical protein n=1 Tax=... 87 8e-16
UniRef100_UPI0000ECA7EA U4/U6 small nuclear ribonucleoprotein Pr... 87 8e-16
UniRef100_Q4S349 Chromosome 4 SCAF14752, whole genome shotgun se... 87 8e-16
UniRef100_Q8Z0R1 WD-40 repeat protein n=1 Tax=Nostoc sp. PCC 712... 87 8e-16
UniRef100_B1X1Y9 WD-repeat protein n=1 Tax=Cyanothece sp. ATCC 5... 87 8e-16
UniRef100_B7G7B5 PolyAdenylation factor subunit 1 (Fragment) n=1... 87 8e-16
UniRef100_B4KNA2 GI20797 n=1 Tax=Drosophila mojavensis RepID=B4K... 87 8e-16
UniRef100_B8MUS1 G-protein beta WD-40 repeats containing protein... 87 8e-16
UniRef100_Q115C0 Serine/threonine protein kinase with WD40 repea... 79 8e-16
UniRef100_B8MI40 Wd40 protein, putative n=1 Tax=Talaromyces stip... 77 1e-15
UniRef100_B0C6X5 WD-repeat protein n=1 Tax=Acaryochloris marina ... 82 1e-15
UniRef100_UPI00004F5540 PREDICTED: similar to TAF5-like RNA poly... 86 1e-15
UniRef100_Q4SVI5 Chromosome 18 SCAF13757, whole genome shotgun s... 86 1e-15
UniRef100_Q7NM62 WD-repeat protein n=1 Tax=Gloeobacter violaceus... 86 1e-15
UniRef100_B2IVX2 WD-40 repeat protein n=1 Tax=Nostoc punctiforme... 86 1e-15
UniRef100_B4VN22 Putative uncharacterized protein n=1 Tax=Microc... 86 1e-15
UniRef100_B9GM11 Predicted protein (Fragment) n=1 Tax=Populus tr... 86 1e-15
UniRef100_B8A0D2 Putative uncharacterized protein n=1 Tax=Zea ma... 86 1e-15
UniRef100_B4HN17 GM20485 n=1 Tax=Drosophila sechellia RepID=B4HN... 86 1e-15
UniRef100_A0EG79 Chromosome undetermined scaffold_94, whole geno... 86 1e-15
UniRef100_Q8YN14 WD-repeat protein n=1 Tax=Nostoc sp. PCC 7120 R... 79 1e-15
UniRef100_A7EJJ2 Putative uncharacterized protein n=1 Tax=Sclero... 86 1e-15
UniRef100_A0DJA3 Chromosome undetermined scaffold_527, whole gen... 82 1e-15
UniRef100_A0YLR0 WD-repeat protein n=1 Tax=Lyngbya sp. PCC 8106 ... 83 1e-15
UniRef100_UPI0000F2B3AD PREDICTED: similar to PRP4 pre-mRNA proc... 86 1e-15
UniRef100_UPI00005EB6BF PREDICTED: similar to PRP4 pre-mRNA proc... 86 1e-15
UniRef100_UPI00001C924B PRP4 pre-mRNA processing factor 4 homolo... 86 1e-15
UniRef100_B4VLP3 Putative uncharacterized protein n=1 Tax=Microc... 86 1e-15
UniRef100_Q28YL4 GA20529 n=1 Tax=Drosophila pseudoobscura pseudo... 86 1e-15
UniRef100_C7G084 WD40 repeat-containing protein n=1 Tax=Dictyost... 86 1e-15
UniRef100_B4QAP5 GD25948 n=1 Tax=Drosophila simulans RepID=B4QAP... 86 1e-15
UniRef100_B4P7N1 GE13065 n=1 Tax=Drosophila yakuba RepID=B4P7N1_... 86 1e-15