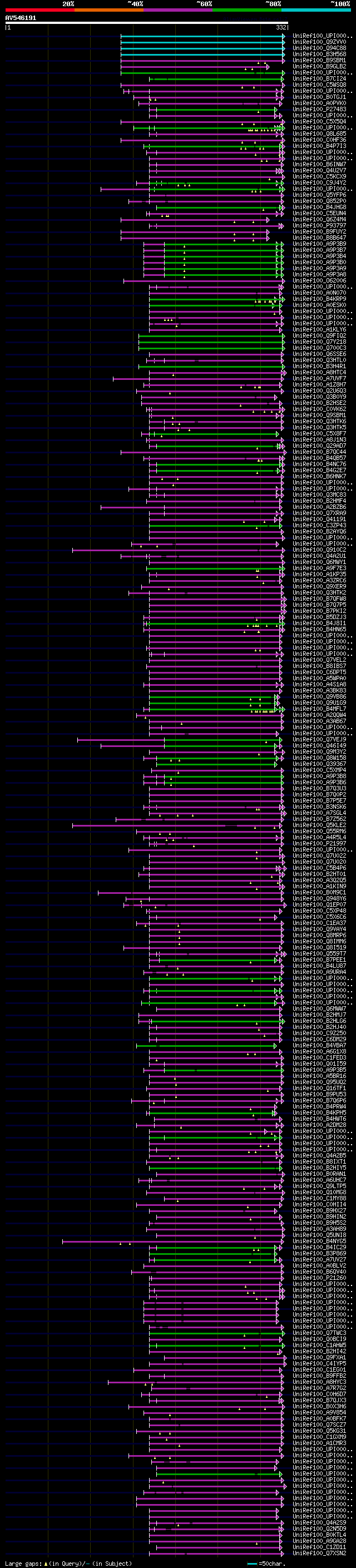
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV546191 RZL10a07F
(332 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_UPI0000163158 emb1011 (embryo defective 1011) n=1 Tax=... 139 1e-31
UniRef100_Q9ZVV0 T5A14.5 protein n=1 Tax=Arabidopsis thaliana Re... 139 1e-31
UniRef100_Q94C88 Putative uncharacterized protein At1g55540 (Fra... 139 1e-31
UniRef100_B3H568 Uncharacterized protein At1g55540.2 n=1 Tax=Ara... 139 1e-31
UniRef100_B9SBM1 Nuclear pore complex protein nup153, putative n... 77 8e-13
UniRef100_B9GLB2 Predicted protein n=1 Tax=Populus trichocarpa R... 67 8e-10
UniRef100_UPI00019836A4 PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_B7CI24 Endo/excinuclease domain protein n=1 Tax=Burkho... 63 1e-08
UniRef100_C5WSQ8 Putative uncharacterized protein Sb01g042700 n=... 62 2e-08
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 62 3e-08
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 62 3e-08
UniRef100_A0PVK0 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 61 3e-08
UniRef100_P27483 Glycine-rich cell wall structural protein n=1 T... 61 3e-08
UniRef100_UPI0000D5650F PREDICTED: similar to GCR(ich) CG5812-PA... 61 4e-08
UniRef100_C5X5Q4 Putative uncharacterized protein Sb02g043330 n=... 61 4e-08
UniRef100_UPI000186F1F7 heavy-chain filboin, putative n=1 Tax=Pe... 60 6e-08
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 60 6e-08
UniRef100_C0HF36 Putative uncharacterized protein n=1 Tax=Zea ma... 60 6e-08
UniRef100_B4P7I3 GE13038 n=1 Tax=Drosophila yakuba RepID=B4P7I3_... 60 6e-08
UniRef100_UPI0001BB95DB predicted protein n=1 Tax=Acinetobacter ... 60 8e-08
UniRef100_UPI0001B7A53E UPI0001B7A53E related cluster n=1 Tax=Ra... 60 8e-08
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 60 8e-08
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 60 8e-08
UniRef100_C5KCX9 Putative uncharacterized protein n=1 Tax=Perkin... 60 8e-08
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 60 8e-08
UniRef100_UPI0000DB7618 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_Q5YFP6 Putative uncharacterized protein n=1 Tax=Singap... 60 1e-07
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 60 1e-07
UniRef100_B4JHG8 GH18631 n=1 Tax=Drosophila grimshawi RepID=B4JH... 60 1e-07
UniRef100_C5EUN4 Predicted protein n=1 Tax=Clostridiales bacteri... 59 1e-07
UniRef100_Q6Z4M4 Os07g0685400 protein n=1 Tax=Oryza sativa Japon... 59 1e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 59 1e-07
UniRef100_B9FUY2 Putative uncharacterized protein n=1 Tax=Oryza ... 59 1e-07
UniRef100_B8B647 Putative uncharacterized protein n=1 Tax=Oryza ... 59 1e-07
UniRef100_A9P3B9 Anther-specific proline rich protein n=1 Tax=Br... 59 1e-07
UniRef100_A9P3B7 Anther-specific proline rich protein n=1 Tax=Br... 59 1e-07
UniRef100_A9P3B4 Anther-specific proline rich protein n=1 Tax=Br... 59 1e-07
UniRef100_A9P3B0 Anther-specific proline rich protein n=1 Tax=Br... 59 1e-07
UniRef100_A9P3A9 Anther-specific proline rich protein n=4 Tax=Br... 59 1e-07
UniRef100_A9P3A8 Anther-specific proline rich protein n=1 Tax=Br... 59 1e-07
UniRef100_O62006 Intermediate filament protein C2 n=1 Tax=Branch... 59 1e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 59 2e-07
UniRef100_A0N070 Glycine-rich protein n=1 Tax=Gossypium hirsutum... 59 2e-07
UniRef100_B4KRP9 GI19648 n=1 Tax=Drosophila mojavensis RepID=B4K... 59 2e-07
UniRef100_A0ESK0 Glycine/tyrosine-rich eggshell protein n=1 Tax=... 59 2e-07
UniRef100_UPI0001AF6E17 PE-PGRS family protein n=1 Tax=Mycobacte... 59 2e-07
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_A1KLY6 PE-PGRS family protein n=2 Tax=Mycobacterium bo... 59 2e-07
UniRef100_Q9FIQ2 Genomic DNA, chromosome 5, P1 clone:MZA15 n=1 T... 59 2e-07
UniRef100_Q7Y218 Putative uncharacterized protein At5g46730 n=1 ... 59 2e-07
UniRef100_Q700C3 At5g46730 (Fragment) n=1 Tax=Arabidopsis thalia... 59 2e-07
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 59 2e-07
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 59 2e-07
UniRef100_B3H4R1 Uncharacterized protein At5g46730.2 n=1 Tax=Ara... 59 2e-07
UniRef100_A8HTC4 Predicted protein n=1 Tax=Chlamydomonas reinhar... 59 2e-07
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 59 2e-07
UniRef100_A1Z8H7 Cuticular protein 47Ef, isoform A n=2 Tax=Droso... 59 2e-07
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 59 2e-07
UniRef100_Q3B0Y9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 58 3e-07
UniRef100_B2HSE2 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 58 3e-07
UniRef100_C0VK62 Host factor Hfq n=1 Tax=Acinetobacter sp. ATCC ... 58 3e-07
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 58 3e-07
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 58 3e-07
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 58 3e-07
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 58 3e-07
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 58 3e-07
UniRef100_Q29AD7 GA19149 n=1 Tax=Drosophila pseudoobscura pseudo... 58 3e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 58 3e-07
UniRef100_B4QB57 GD25930 n=1 Tax=Drosophila simulans RepID=B4QB5... 58 3e-07
UniRef100_B4NC76 GK25807 n=1 Tax=Drosophila willistoni RepID=B4N... 58 3e-07
UniRef100_B4G2E7 GL23896 n=1 Tax=Drosophila persimilis RepID=B4G... 58 3e-07
UniRef100_B6HNK7 Pc21g05120 protein n=1 Tax=Penicillium chrysoge... 58 3e-07
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 58 4e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 58 4e-07
UniRef100_Q3MC83 Putative uncharacterized protein n=1 Tax=Anabae... 58 4e-07
UniRef100_B2HMF4 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 58 4e-07
UniRef100_A2BZB6 RNA-binding region RNP-1 (RNA recognition motif... 58 4e-07
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 58 4e-07
UniRef100_Q41191 Glycine-rich protein (Fragment) n=1 Tax=Arabido... 58 4e-07
UniRef100_C3ZP43 Putative uncharacterized protein n=1 Tax=Branch... 58 4e-07
UniRef100_B2AYQ6 Predicted CDS Pa_1_11890 n=1 Tax=Podospora anse... 58 4e-07
UniRef100_UPI00019019B5 hypothetical protein MtubE_16994 n=1 Tax... 57 5e-07
UniRef100_UPI0000D9B284 PREDICTED: similar to formin 3 CG33556-P... 57 5e-07
UniRef100_Q910C2 Adult keratin RAK (Fragment) n=1 Tax=Rana cates... 57 5e-07
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 57 5e-07
UniRef100_Q6MWY1 PE-PGRS FAMILY PROTEIN (Fragment) n=1 Tax=Mycob... 57 5e-07
UniRef100_A9F7E3 PE-PGRS family protein n=1 Tax=Sorangium cellul... 57 5e-07
UniRef100_A1KP35 PE_PGRS50 protein n=2 Tax=Mycobacterium bovis B... 57 5e-07
UniRef100_A3ZRC6 Putative uncharacterized protein n=1 Tax=Blasto... 57 5e-07
UniRef100_Q9XER9 Putative transcription factor n=1 Tax=Arabidops... 57 5e-07
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 57 5e-07
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 57 5e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 57 5e-07
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 57 5e-07
UniRef100_B5DZJ3 GA24283 n=1 Tax=Drosophila pseudoobscura pseudo... 57 5e-07
UniRef100_B4J8I1 GH21940 n=1 Tax=Drosophila grimshawi RepID=B4J8... 57 5e-07
UniRef100_B4HN65 GM20461 n=1 Tax=Drosophila sechellia RepID=B4HN... 57 5e-07
UniRef100_UPI0001B44D1F hypothetical protein MtubK_12020 n=1 Tax... 57 6e-07
UniRef100_UPI0001B44BAF hypothetical protein MtubKZN_09322 n=1 T... 57 6e-07
UniRef100_UPI00015B5E40 PREDICTED: similar to CG4038-PA n=1 Tax=... 57 6e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_Q7VEL2 Putative uncharacterized protein PE_PGRS37 n=1 ... 57 6e-07
UniRef100_B8IBS7 Loricrin-like protein n=1 Tax=Methylobacterium ... 57 6e-07
UniRef100_C6DPT5 PE-PGRS family protein n=5 Tax=Mycobacterium tu... 57 6e-07
UniRef100_A5WPA0 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 57 6e-07
UniRef100_A4S1A8 Predicted protein n=1 Tax=Ostreococcus lucimari... 57 6e-07
UniRef100_A3BK83 Putative uncharacterized protein n=1 Tax=Oryza ... 57 6e-07
UniRef100_Q9VB86 TweedleT n=1 Tax=Drosophila melanogaster RepID=... 57 6e-07
UniRef100_Q9U1G9 Gly-rich protein n=1 Tax=Drosophila melanogaste... 57 6e-07
UniRef100_B4MFL7 GJ15014 n=1 Tax=Drosophila virilis RepID=B4MFL7... 57 6e-07
UniRef100_A2QQW4 Contig An08c0110, complete genome n=1 Tax=Asper... 57 6e-07
UniRef100_A3AB67 Formin-like protein 16 n=1 Tax=Oryza sativa Jap... 57 6e-07
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 57 8e-07
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 57 8e-07
UniRef100_Q7VEJ9 RNA-binding protein, RRM domain n=1 Tax=Prochlo... 57 8e-07
UniRef100_Q46I49 RNA-binding region RNP-1 (RNA recognition motif... 57 8e-07
UniRef100_Q9M3Y2 Glycine-rich protein n=1 Tax=Triticum aestivum ... 57 8e-07
UniRef100_Q8W158 Anther-specific proline-rich protein n=1 Tax=Br... 57 8e-07
UniRef100_Q39367 Glycine-rich protein (Fragment) n=1 Tax=Brassic... 57 8e-07
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 57 8e-07
UniRef100_A9P3B8 Anther-specific proline rich protein n=1 Tax=Br... 57 8e-07
UniRef100_A9P3B6 Anther-specific proline rich protein n=1 Tax=Br... 57 8e-07
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_B3NSK6 GG22683 n=1 Tax=Drosophila erecta RepID=B3NSK6_... 57 8e-07
UniRef100_A7SGL4 Predicted protein n=1 Tax=Nematostella vectensi... 57 8e-07
UniRef100_B7Z562 cDNA FLJ59041 n=1 Tax=Homo sapiens RepID=B7Z562... 57 8e-07
UniRef100_Q5KLE2 Putative uncharacterized protein n=1 Tax=Filoba... 57 8e-07
UniRef100_Q55RM6 Putative uncharacterized protein n=1 Tax=Filoba... 57 8e-07
UniRef100_A4R5L4 Putative uncharacterized protein n=1 Tax=Magnap... 57 8e-07
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 57 8e-07
UniRef100_UPI000185F332 hypothetical protein BRAFLDRAFT_199348 n... 56 1e-06
UniRef100_Q7U022 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 56 1e-06
UniRef100_Q7U020 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 56 1e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 56 1e-06
UniRef100_B2HT01 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 56 1e-06
UniRef100_A3Q2Q5 Putative uncharacterized protein n=1 Tax=Mycoba... 56 1e-06
UniRef100_A1KIN9 PE-PGRS family protein n=2 Tax=Mycobacterium bo... 56 1e-06
UniRef100_B0M9C1 Putative uncharacterized protein n=1 Tax=Anaero... 56 1e-06
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 56 1e-06
UniRef100_Q1EP07 Putative uncharacterized protein n=1 Tax=Musa a... 56 1e-06
UniRef100_C5XP48 Putative uncharacterized protein Sb03g005056 (F... 56 1e-06
UniRef100_C5X6C6 Putative uncharacterized protein Sb02g043650 n=... 56 1e-06
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 1e-06
UniRef100_Q9VAY4 CG5514, isoform A n=1 Tax=Drosophila melanogast... 56 1e-06
UniRef100_Q8MRP6 GH07623p n=1 Tax=Drosophila melanogaster RepID=... 56 1e-06
UniRef100_Q8IMM6 CG5514, isoform B n=1 Tax=Drosophila melanogast... 56 1e-06
UniRef100_Q8I519 Erythrocyte membrane protein 1, PfEMP1 n=1 Tax=... 56 1e-06
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 56 1e-06
UniRef100_B7PEE1 Nucleolar protein, putative (Fragment) n=1 Tax=... 56 1e-06
UniRef100_B4LU87 GJ17267 n=1 Tax=Drosophila virilis RepID=B4LU87... 56 1e-06
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 56 1e-06
UniRef100_UPI00019832CC PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI00017F0C2D PREDICTED: similar to keratin 3 n=1 Tax=... 56 1e-06
UniRef100_UPI000155BA60 PREDICTED: similar to hCG2029577, partia... 56 1e-06
UniRef100_UPI00004D4BCD Keratin, type II cytoskeletal 5 (Cytoker... 56 1e-06
UniRef100_Q6MWW7 PE-PGRS FAMILY PROTEIN (Fragment) n=1 Tax=Mycob... 56 1e-06
UniRef100_B2HMJ7 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 56 1e-06
UniRef100_B2HLG6 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 56 1e-06
UniRef100_B2HJ40 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 56 1e-06
UniRef100_C9Z250 Putative translation initiation/elongation fact... 56 1e-06
UniRef100_C6DM29 Predicted protein n=1 Tax=Mycobacterium tubercu... 56 1e-06
UniRef100_B4VBA7 Putative uncharacterized protein n=1 Tax=Strept... 56 1e-06
UniRef100_A6G1X8 Single-stranded DNA-binding protein n=1 Tax=Ple... 56 1e-06
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 1e-06
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 56 1e-06
UniRef100_A9P3B5 Anther-specific proline rich protein n=1 Tax=Br... 56 1e-06
UniRef100_A5BR16 Putative uncharacterized protein n=1 Tax=Vitis ... 56 1e-06
UniRef100_Q95UQ2 Subtilisin-like protein n=1 Tax=Toxoplasma gond... 56 1e-06
UniRef100_Q16TF1 Fibrillarin (Fragment) n=1 Tax=Aedes aegypti Re... 56 1e-06
UniRef100_B9PU53 Subtilase family protein n=1 Tax=Toxoplasma gon... 56 1e-06
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B4PRW4 GE23709 n=1 Tax=Drosophila yakuba RepID=B4PRW4_... 56 1e-06
UniRef100_B4KPH5 GI18675 n=1 Tax=Drosophila mojavensis RepID=B4K... 56 1e-06
UniRef100_B4HWT6 GM11565 n=1 Tax=Drosophila sechellia RepID=B4HW... 56 1e-06
UniRef100_A2DM28 Diaphanous, putative n=1 Tax=Trichomonas vagina... 56 1e-06
UniRef100_UPI0001BBAA33 predicted protein n=1 Tax=Acinetobacter ... 55 2e-06
UniRef100_UPI00017962B2 PREDICTED: similar to keratin 3 n=1 Tax=... 55 2e-06
UniRef100_UPI0001792863 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI00015B52EC PREDICTED: similar to ENSANGP00000014755... 55 2e-06
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 55 2e-06
UniRef100_B8IXT1 Loricrin-like protein n=1 Tax=Methylobacterium ... 55 2e-06
UniRef100_B2HIY5 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 55 2e-06
UniRef100_B0RAN1 Putative integral membrane protein n=1 Tax=Clav... 55 2e-06
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 55 2e-06
UniRef100_Q9LTP5 Genomic DNA, chromosome 3, P1 clone: MQC12 n=1 ... 55 2e-06
UniRef100_Q10MG8 Retrotransposon protein, putative, Ty3-gypsy su... 55 2e-06
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 2e-06
UniRef100_C0HII4 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_B9HX27 Predicted protein n=1 Tax=Populus trichocarpa R... 55 2e-06
UniRef100_B9HIN2 Predicted protein n=1 Tax=Populus trichocarpa R... 55 2e-06
UniRef100_B9H5S2 Predicted protein n=1 Tax=Populus trichocarpa R... 55 2e-06
UniRef100_A3AH89 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_Q5UNI8 Dragline silk spidroin 1 (Fragment) n=1 Tax=Nep... 55 2e-06
UniRef100_B4NYG5 GE14785 n=1 Tax=Drosophila yakuba RepID=B4NYG5_... 55 2e-06
UniRef100_B4IC29 GM10360 n=1 Tax=Drosophila sechellia RepID=B4IC... 55 2e-06
UniRef100_B3P869 GG11520 n=1 Tax=Drosophila erecta RepID=B3P869_... 55 2e-06
UniRef100_A7UV27 Cuticular protein 133, RR-1 family (AGAP009872-... 55 2e-06
UniRef100_A0BLV2 Chromosome undetermined scaffold_115, whole gen... 55 2e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 55 2e-06
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 55 2e-06
UniRef100_UPI0001AF6BC1 hypothetical protein MkanA1_22809 n=1 Ta... 55 2e-06
UniRef100_UPI00019856A2 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI000185F331 hypothetical protein BRAFLDRAFT_64022 n=... 55 2e-06
UniRef100_UPI00017C32F4 PREDICTED: similar to Histone-lysine N-m... 55 2e-06
UniRef100_UPI00005A499E PREDICTED: similar to CG40351-PA.3 n=1 T... 55 2e-06
UniRef100_UPI000184A044 SET domain-containing protein 1B n=1 Tax... 55 2e-06
UniRef100_UPI000179DAA8 UPI000179DAA8 related cluster n=1 Tax=Bo... 55 2e-06
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 55 2e-06
UniRef100_Q7TWC3 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 55 2e-06
UniRef100_Q0BCI9 Putative uncharacterized protein n=1 Tax=Burkho... 55 2e-06
UniRef100_C1AHW5 PE-PGRS family protein n=1 Tax=Mycobacterium bo... 55 2e-06
UniRef100_B2HI42 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 55 2e-06
UniRef100_Q9FXA1 AT1G49750 protein n=1 Tax=Arabidopsis thaliana ... 55 2e-06
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_C1EG01 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 2e-06
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 55 2e-06
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 55 2e-06
UniRef100_C0H6D7 Putative cuticle protein n=1 Tax=Bombyx mori Re... 55 2e-06
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B0X3H6 Putative uncharacterized protein n=1 Tax=Culex ... 55 2e-06
UniRef100_A9V854 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 2e-06
UniRef100_A0BFK7 Chromosome undetermined scaffold_104, whole gen... 55 2e-06
UniRef100_Q7SCZ7 WASP-interacting protein-like protein vrp1p n=1... 55 2e-06
UniRef100_Q5KG31 Putative uncharacterized protein n=1 Tax=Filoba... 55 2e-06
UniRef100_C1GXM9 Cytokinesis protein sepA n=1 Tax=Paracoccidioid... 55 2e-06
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 55 2e-06
UniRef100_UPI00019856A3 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI00019261C6 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 55 3e-06
UniRef100_UPI000155434A PREDICTED: similar to olfactory receptor... 55 3e-06
UniRef100_UPI0000F2E2A9 PREDICTED: similar to keratin 3 n=1 Tax=... 55 3e-06
UniRef100_UPI0000E4922C PREDICTED: similar to Enabled homolog (D... 55 3e-06
UniRef100_UPI00006A0D59 UPI00006A0D59 related cluster n=1 Tax=Xe... 55 3e-06
UniRef100_UPI0001AE6B4C SET domain-containing protein 1B n=1 Tax... 55 3e-06
UniRef100_UPI00015DF9DB Proline-rich protein 12. n=1 Tax=Homo sa... 55 3e-06
UniRef100_UPI0001596889 proline rich 12 n=1 Tax=Homo sapiens Rep... 55 3e-06
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 55 3e-06
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 55 3e-06
UniRef100_B0KTL4 Putative uncharacterized protein n=1 Tax=Pseudo... 55 3e-06
UniRef100_A9GA28 PE-PGRS family protein n=1 Tax=Sorangium cellul... 55 3e-06
UniRef100_C1ZD11 Putative uncharacterized protein n=1 Tax=Planct... 55 3e-06
UniRef100_Q7XSN2 OSJNBb0028M18.5 protein n=1 Tax=Oryza sativa Ja... 55 3e-06
UniRef100_Q7XHB6 Transposon protein, putative, CACTA, En/Spm sub... 55 3e-06
UniRef100_Q6Z498 Os07g0511100 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 55 3e-06
UniRef100_Q41187 Glycine-rich protein (Fragment) n=1 Tax=Arabido... 55 3e-06
UniRef100_Q2QWV3 Transposon protein, putative, CACTA, En/Spm sub... 55 3e-06
UniRef100_Q01MU7 OSIGBa0102O13.9 protein n=1 Tax=Oryza sativa Re... 55 3e-06
UniRef100_C4J5K1 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_C1MMP7 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 3e-06
UniRef100_B9SS24 Glycine-rich cell wall structural protein 1.8, ... 55 3e-06
UniRef100_B9MU29 Predicted protein n=1 Tax=Populus trichocarpa R... 55 3e-06
UniRef100_B6U1N0 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 55 3e-06
UniRef100_Q0IGW9 LP21747p n=1 Tax=Drosophila melanogaster RepID=... 55 3e-06
UniRef100_O46171 Spidroin 1 (Fragment) n=1 Tax=Nephila clavipes ... 55 3e-06
UniRef100_O44357 Abductin n=1 Tax=Argopecten irradians RepID=O44... 55 3e-06
UniRef100_B4M120 GJ23081 n=1 Tax=Drosophila virilis RepID=B4M120... 55 3e-06
UniRef100_A1Z8H8 Cuticular protein 47Ef, isoform B n=1 Tax=Droso... 55 3e-06
UniRef100_A6NFN6 Putative uncharacterized protein PRR12 n=1 Tax=... 55 3e-06
UniRef100_C5G899 Predicted protein n=1 Tax=Ajellomyces dermatiti... 55 3e-06
UniRef100_A8NQG1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 55 3e-06
UniRef100_Q8CFT2-2 Isoform 2 of Histone-lysine N-methyltransfera... 55 3e-06
UniRef100_Q8CFT2 Histone-lysine N-methyltransferase SETD1B n=2 T... 55 3e-06
UniRef100_Q9UPS6 Histone-lysine N-methyltransferase SETD1B n=1 T... 55 3e-06
UniRef100_Q9ULL5-2 Isoform 2 of Proline-rich protein 12 n=1 Tax=... 55 3e-06
UniRef100_Q9ULL5 Proline-rich protein 12 n=1 Tax=Homo sapiens Re... 55 3e-06
UniRef100_B1VYN5 Translation initiation factor IF-2 n=1 Tax=Stre... 55 3e-06
UniRef100_A4X4N7 Translation initiation factor IF-2 n=1 Tax=Sali... 55 3e-06
UniRef100_Q9C0D6 FH2 domain-containing protein 1 n=1 Tax=Homo sa... 55 3e-06
UniRef100_Q54IP0 DnaJ homolog subfamily C member 7 homolog n=1 T... 55 3e-06
UniRef100_UPI0001861057 hypothetical protein BRAFLDRAFT_119668 n... 54 4e-06
UniRef100_UPI000175F433 PREDICTED: similar to formin-like 1 n=1 ... 54 4e-06
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI00015B42C5 PREDICTED: similar to conserved hypothet... 54 4e-06
UniRef100_UPI00015551C8 PREDICTED: similar to seven transmembran... 54 4e-06
UniRef100_UPI0000E82531 PREDICTED: similar to Krt6b protein n=1 ... 54 4e-06
UniRef100_UPI0001A2D69A UPI0001A2D69A related cluster n=1 Tax=Da... 54 4e-06
UniRef100_UPI0000DC1294 UPI0000DC1294 related cluster n=1 Tax=Ra... 54 4e-06
UniRef100_UPI0000EC9E20 UPI0000EC9E20 related cluster n=1 Tax=Ga... 54 4e-06
UniRef100_Q6DD43 LOC446221 protein (Fragment) n=1 Tax=Xenopus la... 54 4e-06
UniRef100_Q5ID03 Enabled protein n=1 Tax=Xenopus laevis RepID=Q5... 54 4e-06
UniRef100_Q4V859 LOC446221 protein n=1 Tax=Xenopus laevis RepID=... 54 4e-06
UniRef100_B2GUG8 LOC100158585 protein n=1 Tax=Xenopus (Silurana)... 54 4e-06
UniRef100_A1L2T3 LOC100036944 protein n=1 Tax=Xenopus laevis Rep... 54 4e-06
UniRef100_Q8QKX8 EsV-1-144 n=1 Tax=Ectocarpus siliculosus virus ... 54 4e-06
UniRef100_A4FTH0 Putative uncharacterized protein n=1 Tax=Cyprin... 54 4e-06
UniRef100_A3QMN0 ORF62 n=1 Tax=Cyprinid herpesvirus 3 RepID=A3QM... 54 4e-06
UniRef100_Q7U160 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 54 4e-06
UniRef100_Q7TWB8 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 54 4e-06
UniRef100_Q79FV7 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_Q6MWW9 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_C1AHX0 PE-PGRS family protein n=1 Tax=Mycobacterium bo... 54 4e-06
UniRef100_B4EAT8 Single-stranded DNA-binding protein n=1 Tax=Bur... 54 4e-06
UniRef100_B2HM19 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 54 4e-06
UniRef100_B2HDQ3 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 54 4e-06
UniRef100_A9GB80 Putative secreted protein n=1 Tax=Sorangium cel... 54 4e-06
UniRef100_A9F6B6 Putative uncharacterized protein n=1 Tax=Sorang... 54 4e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 54 4e-06
UniRef100_A5U8I2 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_A1UJA1 PE-PGRS family protein n=2 Tax=Mycobacterium Re... 54 4e-06
UniRef100_A1KPJ3 PE-PGRS family protein n=1 Tax=Mycobacterium bo... 54 4e-06
UniRef100_A1KGW5 PE-PGRS family protein n=2 Tax=Mycobacterium bo... 54 4e-06
UniRef100_Q8VKN7 Putative uncharacterized protein n=1 Tax=Mycoba... 54 4e-06
UniRef100_Q8VIZ0 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_Q7D974 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_D0C648 Predicted protein n=1 Tax=Acinetobacter baumann... 54 4e-06
UniRef100_B0V7B2 Host factor I for bacteriophage Q beta replicat... 54 4e-06
UniRef100_C9NAA4 Putative uncharacterized protein n=1 Tax=Strept... 54 4e-06
UniRef100_C6DVD8 PE-PGRS family protein n=2 Tax=Mycobacterium tu... 54 4e-06
UniRef100_C6DM26 Predicted protein n=1 Tax=Mycobacterium tubercu... 54 4e-06
UniRef100_A8E485 HrpW n=1 Tax=Pseudomonas viridiflava RepID=A8E4... 54 4e-06
UniRef100_A5WT78 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_A5WKK2 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 4e-06
UniRef100_Q8H797 Putative uncharacterized protein (Fragment) n=1... 54 4e-06
UniRef100_Q8GTL0 Putative glycine-rich cell wall protein n=2 Tax... 54 4e-06
UniRef100_Q7XES4 Retrotransposon protein, putative, unclassified... 54 4e-06
UniRef100_Q2QVX3 Transposon protein, putative, CACTA, En/Spm sub... 54 4e-06
UniRef100_C7J5E4 Os07g0688700 protein (Fragment) n=1 Tax=Oryza s... 54 4e-06
UniRef100_C5XD76 Putative uncharacterized protein Sb02g038276 (F... 54 4e-06
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 4e-06
UniRef100_B9RS81 Serine-threonine protein kinase, plant-type, pu... 54 4e-06
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 54 4e-06
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 54 4e-06
UniRef100_A8J3Q3 Glyoxal or galactose oxidase n=1 Tax=Chlamydomo... 54 4e-06
UniRef100_A8ITW8 Dicer-like protein n=1 Tax=Chlamydomonas reinha... 54 4e-06
UniRef100_A5AKR0 Putative uncharacterized protein n=1 Tax=Vitis ... 54 4e-06
UniRef100_A2YQ51 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_A2YLR2 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_O44356 Abductin n=1 Tax=Argopecten irradians RepID=O44... 54 4e-06
UniRef100_B4IGY1 GM16364 n=1 Tax=Drosophila sechellia RepID=B4IG... 54 4e-06
UniRef100_Q75DI6 ABR038Cp n=1 Tax=Eremothecium gossypii RepID=Q7... 54 4e-06
UniRef100_C7YU47 Putative uncharacterized protein n=1 Tax=Nectri... 54 4e-06
UniRef100_UPI0001AF6ED8 hypothetical protein MkanA1_26432 n=1 Ta... 54 5e-06
UniRef100_UPI000198414B PREDICTED: hypothetical protein isoform ... 54 5e-06
UniRef100_UPI000198414A PREDICTED: hypothetical protein isoform ... 54 5e-06
UniRef100_UPI000194C8B0 PREDICTED: dishevelled associated activa... 54 5e-06
UniRef100_UPI000194C8AE PREDICTED: dishevelled associated activa... 54 5e-06
UniRef100_UPI000186E9B0 conserved hypothetical protein n=1 Tax=P... 54 5e-06
UniRef100_UPI000186CD84 hypothetical protein Phum_PHUM040670 n=1... 54 5e-06
UniRef100_UPI00005A1BE2 PREDICTED: similar to keratin 13 isoform... 54 5e-06
UniRef100_UPI0001B7A6BD enabled homolog n=1 Tax=Rattus norvegicu... 54 5e-06
UniRef100_UPI0001B7A6BC enabled homolog n=1 Tax=Rattus norvegicu... 54 5e-06
UniRef100_UPI0001B7A6A7 enabled homolog n=1 Tax=Rattus norvegicu... 54 5e-06
UniRef100_Q7SXQ3 Zgc:66127 n=1 Tax=Danio rerio RepID=Q7SXQ3_DANRE 54 5e-06
UniRef100_B7FCD4 Beta-keratin-like protein n=1 Tax=Crocodylus ni... 54 5e-06
UniRef100_Q65553 UL36 n=3 Tax=Bovine herpesvirus 1 RepID=Q65553_... 54 5e-06
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 54 5e-06
UniRef100_Q5XHX3 Enabled homolog (Drosophila) n=1 Tax=Rattus nor... 54 5e-06
UniRef100_Q3UJS2 Putative uncharacterized protein n=1 Tax=Mus mu... 54 5e-06
UniRef100_Q8YQB7 All3916 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 54 5e-06
UniRef100_Q7TWK6 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 54 5e-06
UniRef100_C5C2A2 DEAD/DEAH box helicase domain protein n=1 Tax=B... 54 5e-06
UniRef100_B2HR57 PE-PGRS family protein, PE_PGRS15_2 n=1 Tax=Myc... 54 5e-06
UniRef100_B0UDX5 Loricrin-like protein n=1 Tax=Methylobacterium ... 54 5e-06
UniRef100_A9F183 Putative uncharacterized protein n=1 Tax=Sorang... 54 5e-06
UniRef100_A9B9L1 RNA-binding region RNP-1 (RNA recognition motif... 54 5e-06
UniRef100_A8LE82 2-oxoglutarate dehydrogenase E2 component n=1 T... 54 5e-06
UniRef100_A1UN99 Putative uncharacterized protein n=2 Tax=Mycoba... 54 5e-06
UniRef100_A1T4J8 Collagen triple helix repeat n=1 Tax=Mycobacter... 54 5e-06
UniRef100_A1KP78 PE-PGRS family protein n=2 Tax=Mycobacterium bo... 54 5e-06
UniRef100_A0PVL1 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 54 5e-06
UniRef100_Q8VJ15 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 54 5e-06
UniRef100_C7NET7 Putative uncharacterized protein n=1 Tax=Kytoco... 54 5e-06
UniRef100_C7JH29 Putative uncharacterized protein n=8 Tax=Acetob... 54 5e-06
UniRef100_C6WDK8 Translation initiation factor IF-2 n=1 Tax=Acti... 54 5e-06
UniRef100_C6DLC8 PE-PGRS family protein n=2 Tax=Mycobacterium tu... 54 5e-06
UniRef100_C4ZIL1 Putative uncharacterized protein n=1 Tax=Thauer... 54 5e-06
UniRef100_C2B0M4 Putative uncharacterized protein n=1 Tax=Citrob... 54 5e-06
UniRef100_A8RNC4 Putative uncharacterized protein n=1 Tax=Clostr... 54 5e-06
UniRef100_Q9SIH2 Putative uncharacterized protein At2g36120 n=1 ... 54 5e-06
UniRef100_Q9LW52 Genomic DNA, chromosome 3, P1 clone: MLM24 n=1 ... 54 5e-06
UniRef100_Q6QNA3 Proline-rich protein 1 n=1 Tax=Capsicum annuum ... 54 5e-06
UniRef100_Q5VR46 Os01g0180000 protein n=1 Tax=Oryza sativa Japon... 54 5e-06
UniRef100_Q5VR02 Hydroxyproline-rich glycoprotein-like n=1 Tax=O... 54 5e-06
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 54 5e-06
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 54 5e-06
UniRef100_Q41518 Single-stranded nucleic acid binding protein n=... 54 5e-06
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 54 5e-06
UniRef100_Q3ECQ3 Uncharacterized protein At1g54215.1 n=1 Tax=Ara... 54 5e-06
UniRef100_C0PPC0 Putative uncharacterized protein n=1 Tax=Zea ma... 54 5e-06
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 54 5e-06
UniRef100_B8ADK4 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 54 5e-06
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 54 5e-06
UniRef100_A7Z068 LMOD2 protein n=1 Tax=Bos taurus RepID=A7Z068_B... 54 5e-06
UniRef100_Q8SZ23 CG8160 n=1 Tax=Drosophila melanogaster RepID=Q8... 54 5e-06
UniRef100_B7QAD8 Secreted protein, putative (Fragment) n=1 Tax=I... 54 5e-06
UniRef100_B7Q5W9 Secreted protein, putative (Fragment) n=1 Tax=I... 54 5e-06
UniRef100_B7PE98 Glycine rich protein, putative (Fragment) n=1 T... 54 5e-06
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 54 5e-06
UniRef100_B4R6E5 GD15691 n=1 Tax=Drosophila simulans RepID=B4R6E... 54 5e-06
UniRef100_B4JWL2 GH23068 n=1 Tax=Drosophila grimshawi RepID=B4JW... 54 5e-06
UniRef100_B4IZJ4 GH14508 n=1 Tax=Drosophila grimshawi RepID=B4IZ... 54 5e-06
UniRef100_B3NJ07 GG16231 n=1 Tax=Drosophila erecta RepID=B3NJ07_... 54 5e-06
UniRef100_A2D765 Putative uncharacterized protein n=1 Tax=Tricho... 54 5e-06
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 54 5e-06
UniRef100_Q875A9 Similar to snoRNP protein gar1 of Schizosacchar... 54 5e-06
UniRef100_Q7SF15 WASP-like pretein las17p n=1 Tax=Neurospora cra... 54 5e-06
UniRef100_Q6AHT0 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 54 5e-06
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 54 5e-06
UniRef100_Q2UUT0 Predicted protein n=1 Tax=Aspergillus oryzae Re... 54 5e-06
UniRef100_Q2U304 Predicted protein n=1 Tax=Aspergillus oryzae Re... 54 5e-06
UniRef100_C8VGJ1 Putative uncharacterized protein n=2 Tax=Emeric... 54 5e-06
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 54 5e-06
UniRef100_C7YMP5 Putative uncharacterized protein n=1 Tax=Nectri... 54 5e-06
UniRef100_B2VLB0 Predicted CDS Pa_5_5520 n=1 Tax=Podospora anser... 54 5e-06
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 54 5e-06
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 54 5e-06
UniRef100_P22509 rRNA 2'-O-methyltransferase fibrillarin n=1 Tax... 54 5e-06
UniRef100_P35550 rRNA 2'-O-methyltransferase fibrillarin n=1 Tax... 54 5e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 54 5e-06
UniRef100_UPI0001B44CDC hypothetical protein MtubK_14878 n=1 Tax... 54 7e-06
UniRef100_UPI0001B44BD1 hypothetical protein MtubKZN_00035 n=1 T... 54 7e-06
UniRef100_UPI0001B44B4F hypothetical protein MtubKZN_12150 n=1 T... 54 7e-06
UniRef100_UPI0001904F99 hypothetical protein MtubT1_15587 n=1 Ta... 54 7e-06
UniRef100_UPI0001903247 hypothetical protein MtubT1_19997 n=1 Ta... 54 7e-06
UniRef100_UPI0001902368 hypothetical protein MtubG1_17864 n=1 Ta... 54 7e-06
UniRef100_UPI0001901A95 hypothetical protein MtubT_06729 n=1 Tax... 54 7e-06
UniRef100_UPI000190197B hypothetical protein MtubT_17880 n=1 Tax... 54 7e-06
UniRef100_UPI000190135F hypothetical protein Mtub0_13120 n=1 Tax... 54 7e-06
UniRef100_UPI0001901005 PE-PGRS family protein n=2 Tax=Mycobacte... 54 7e-06
UniRef100_UPI000186ED02 conserved hypothetical protein n=1 Tax=P... 54 7e-06
UniRef100_UPI000180BC4A PREDICTED: similar to predicted protein ... 54 7e-06
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0000DB6C07 PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI00002210E8 Hypothetical protein CBG07655 n=1 Tax=Ca... 54 7e-06
UniRef100_UPI0000025476 DEAH (Asp-Glu-Ala-His) box polypeptide 9... 54 7e-06
UniRef100_UPI00001EDBAC DEAH (Asp-Glu-Ala-His) box polypeptide 9... 54 7e-06
UniRef100_UPI00016E37F3 UPI00016E37F3 related cluster n=1 Tax=Ta... 54 7e-06
UniRef100_Q8AV12 Adult keratin XAK-C n=1 Tax=Xenopus laevis RepI... 54 7e-06
UniRef100_Q6GM32 MGC84010 protein n=1 Tax=Xenopus laevis RepID=Q... 54 7e-06
UniRef100_B7ZRD4 Ak-c-A protein n=1 Tax=Xenopus laevis RepID=B7Z... 54 7e-06
UniRef100_B0S6Z1 Novel protein similar to vertebrate DEAH (Asp-G... 54 7e-06
UniRef100_B6ID90 DEAH (Asp-Glu-Ala-His) box polypeptide 9 (Fragm... 54 7e-06
UniRef100_Q79FP2 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_B9JQZ2 Putative uncharacterized protein n=1 Tax=Agroba... 54 7e-06
UniRef100_B2HP34 Conserved hypothetical membrane protein, IniB n... 54 7e-06
UniRef100_B2HHZ9 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 54 7e-06
UniRef100_B2HDU1 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 54 7e-06
UniRef100_A9GQI8 Putative uncharacterized protein n=1 Tax=Sorang... 54 7e-06
UniRef100_A5U863 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_A5U2F3 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_A1KPJ8 PE-PGRS family protein n=1 Tax=Mycobacterium bo... 54 7e-06
UniRef100_A0PTJ3 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 54 7e-06
UniRef100_Q8VK17 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_Q8VIZ1 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_C6DTD4 Predicted protein n=1 Tax=Mycobacterium tubercu... 54 7e-06
UniRef100_C6DM25 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_B4D0M3 Putative uncharacterized protein n=1 Tax=Chthon... 54 7e-06
UniRef100_A5WT77 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_A5WSV8 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_P0A691 Uncharacterized PE-PGRS family protein PE_PGRS4... 54 7e-06
UniRef100_A5WMD2 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 54 7e-06
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 54 7e-06
UniRef100_Q5ZD44 Collagen alpha 1 chain-like n=1 Tax=Oryza sativ... 54 7e-06
UniRef100_Q49I33 120 kDa pistil extensin-like protein (Fragment)... 54 7e-06
UniRef100_Q49I32 120 kDa pistil extensin-like protein (Fragment)... 54 7e-06
UniRef100_Q39691 Glycine-rich cell wall protein n=1 Tax=Daucus c... 54 7e-06
UniRef100_Q2QY58 Transposon protein, putative, CACTA, En/Spm sub... 54 7e-06
UniRef100_Q2QQ97 Os12g0502200 protein n=1 Tax=Oryza sativa Japon... 54 7e-06
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 54 7e-06
UniRef100_O24184 Glycine-rich RNA-binding protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_C5X9Y5 Putative uncharacterized protein Sb02g034435 n=... 54 7e-06
UniRef100_C1E9S3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 7e-06
UniRef100_B9SVA0 GRP3S, putative n=1 Tax=Ricinus communis RepID=... 54 7e-06
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 54 7e-06
UniRef100_B9I9M7 Predicted protein n=1 Tax=Populus trichocarpa R... 54 7e-06
UniRef100_A8IZS5 Glycine-rich RNA-binding protein n=1 Tax=Chlamy... 54 7e-06
UniRef100_Q6UDW6 Erythrocyte membrane protein 1 n=1 Tax=Plasmodi... 54 7e-06
UniRef100_B7PK11 Glycine-rich RNA-binding protein, putative (Fra... 54 7e-06
UniRef100_B4I348 GM18116 n=1 Tax=Drosophila sechellia RepID=B4I3... 54 7e-06
UniRef100_B4HXI5 GM15710 n=1 Tax=Drosophila sechellia RepID=B4HX... 54 7e-06
UniRef100_A8X4C8 Putative uncharacterized protein n=1 Tax=Caenor... 54 7e-06
UniRef100_A0NH33 AGAP001510-PA n=1 Tax=Anopheles gambiae RepID=A... 54 7e-06
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 54 7e-06
UniRef100_B2W0T4 Putative uncharacterized protein n=1 Tax=Pyreno... 54 7e-06
UniRef100_A4RL81 Putative uncharacterized protein n=1 Tax=Magnap... 54 7e-06
UniRef100_P19837 Spidroin-1 (Fragment) n=1 Tax=Nephila clavipes ... 54 7e-06
UniRef100_UPI0001B46950 hypothetical protein Mtube_13940 n=1 Tax... 53 9e-06
UniRef100_UPI0001B44C51 PE-PGRS family protein n=1 Tax=Mycobacte... 53 9e-06
UniRef100_UPI0001AF73FF PE-PGRS family protein n=1 Tax=Mycobacte... 53 9e-06
UniRef100_UPI0001A7B366 GRP23 (GLYCINE-RICH PROTEIN 23) n=1 Tax=... 53 9e-06
UniRef100_UPI0001901F49 PE_PGRS family protein n=1 Tax=Mycobacte... 53 9e-06
UniRef100_UPI0001900D62 PE-PGRS family protein n=1 Tax=Mycobacte... 53 9e-06
UniRef100_UPI0001868875 hypothetical protein BRAFLDRAFT_103538 n... 53 9e-06
UniRef100_UPI0001758286 PREDICTED: hypothetical protein n=1 Tax=... 53 9e-06
UniRef100_UPI0001552E25 PREDICTED: hypothetical protein n=1 Tax=... 53 9e-06
UniRef100_UPI0000E460C2 PREDICTED: similar to nucleolin like pro... 53 9e-06
UniRef100_UPI0000DA3EC9 PREDICTED: similar to tumor endothelial ... 53 9e-06
UniRef100_UPI0000DA20D4 PREDICTED: hypothetical protein n=1 Tax=... 53 9e-06
UniRef100_UPI0000DA1F29 PREDICTED: hypothetical protein n=1 Tax=... 53 9e-06
UniRef100_UPI0000DA1BD4 PREDICTED: hypothetical protein n=1 Tax=... 53 9e-06