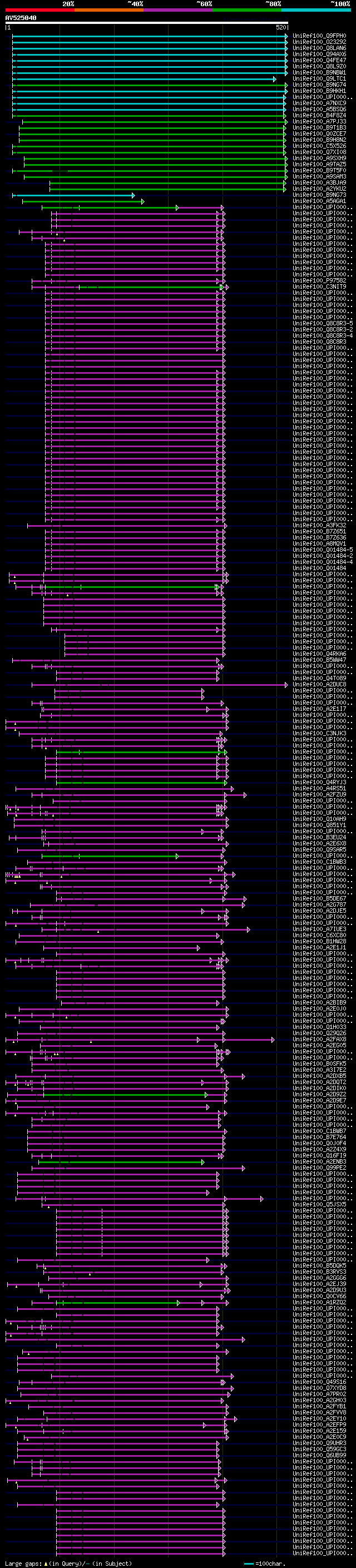
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV525040 APD15f03R
(520 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q9FPH0 AT4g14360 n=1 Tax=Arabidopsis thaliana RepID=Q9... 340 3e-92
UniRef100_O23292 Ankyrin like protein n=1 Tax=Arabidopsis thalia... 340 3e-92
UniRef100_Q8LAN6 Putative uncharacterized protein n=1 Tax=Arabid... 337 3e-91
UniRef100_Q94AX6 AT3g23280/K14B15_17 n=1 Tax=Arabidopsis thalian... 242 1e-62
UniRef100_Q4FE47 At3g23280 n=1 Tax=Arabidopsis thaliana RepID=Q4... 242 1e-62
UniRef100_Q8L9Z0 Putative uncharacterized protein n=1 Tax=Arabid... 239 6e-62
UniRef100_B9NBW1 Predicted protein (Fragment) n=1 Tax=Populus tr... 231 3e-59
UniRef100_Q9LTC1 Similarity to ankyrin like protein n=1 Tax=Arab... 228 2e-58
UniRef100_B9NG74 Predicted protein n=1 Tax=Populus trichocarpa R... 221 2e-56
UniRef100_B9HKH1 Predicted protein (Fragment) n=1 Tax=Populus tr... 221 3e-56
UniRef100_UPI00019834CE PREDICTED: hypothetical protein n=1 Tax=... 219 1e-55
UniRef100_A7NXC9 Chromosome chr5 scaffold_2, whole genome shotgu... 219 1e-55
UniRef100_A5BSQ6 Putative uncharacterized protein n=1 Tax=Vitis ... 219 1e-55
UniRef100_B4F8Z4 Protein binding protein n=1 Tax=Zea mays RepID=... 204 3e-51
UniRef100_A7PJ33 Chromosome chr12 scaffold_18, whole genome shot... 202 1e-50
UniRef100_B9T1B3 Ankyrin repeat domain protein, putative n=1 Tax... 200 4e-50
UniRef100_Q0ZCE7 Auxin-regulated protein-like protein n=1 Tax=Po... 199 9e-50
UniRef100_B9H8N2 Predicted protein n=1 Tax=Populus trichocarpa R... 199 9e-50
UniRef100_C5X526 Putative uncharacterized protein Sb02g010610 n=... 197 4e-49
UniRef100_Q7XI08 Os07g0446100 protein n=1 Tax=Oryza sativa Japon... 189 1e-46
UniRef100_A9SXH9 Predicted protein n=1 Tax=Physcomitrella patens... 179 1e-43
UniRef100_A9TAZ5 Predicted protein n=1 Tax=Physcomitrella patens... 176 9e-43
UniRef100_B9T5F0 S-adenosylmethionine-dependent methyltransferas... 174 4e-42
UniRef100_A9SAM3 Predicted protein n=1 Tax=Physcomitrella patens... 172 1e-41
UniRef100_A3BJA9 Putative uncharacterized protein n=1 Tax=Oryza ... 157 5e-37
UniRef100_A2YKU2 Putative uncharacterized protein n=1 Tax=Oryza ... 157 5e-37
UniRef100_B9NG73 Predicted protein n=1 Tax=Populus trichocarpa R... 96 1e-18
UniRef100_A5AGA1 Putative uncharacterized protein n=1 Tax=Vitis ... 82 2e-14
UniRef100_UPI00001133CF UPI00001133CF related cluster n=1 Tax=un... 76 1e-12
UniRef100_UPI00004D61D9 Ankyrin-2 (Brain ankyrin) (Ankyrin B) (A... 76 1e-12
UniRef100_UPI00004D61D7 Ankyrin-2 (Brain ankyrin) (Ankyrin B) (A... 76 1e-12
UniRef100_UPI00004D61D6 Ankyrin-2 (Brain ankyrin) (Ankyrin B) (A... 76 1e-12
UniRef100_UPI0000E49FCF PREDICTED: similar to ankyrin 2,3/unc44 ... 76 2e-12
UniRef100_UPI0000E46C82 PREDICTED: similar to ankyrin 2,3/unc44,... 76 2e-12
UniRef100_UPI0000E80412 PREDICTED: similar to ankyrin B (440 kDa... 75 2e-12
UniRef100_UPI0000ECC762 Ankyrin 2 n=1 Tax=Gallus gallus RepID=UP... 75 2e-12
UniRef100_UPI000194C4AE PREDICTED: ankyrin 2, neuronal n=1 Tax=T... 75 3e-12
UniRef100_UPI0000F2D69B PREDICTED: similar to ankyrin 2 isoform ... 74 8e-12
UniRef100_UPI0000F2D69A PREDICTED: similar to ankyrin 2 isoform ... 74 8e-12
UniRef100_UPI000155CC79 PREDICTED: similar to ankyrin 2 n=1 Tax=... 73 1e-11
UniRef100_P97582 Ankyrin (Fragment) n=1 Tax=Rattus norvegicus Re... 73 1e-11
UniRef100_C3NIT9 Ankyrin n=1 Tax=Sulfolobus islandicus Y.N.15.51... 73 1e-11
UniRef100_UPI0000DA2162 PREDICTED: similar to ankyrin 2 isoform ... 73 1e-11
UniRef100_UPI0000DA1FD5 PREDICTED: similar to ankyrin 2 isoform ... 73 1e-11
UniRef100_UPI0001B7BC51 Ankyrin n=1 Tax=Rattus norvegicus RepID=... 73 1e-11
UniRef100_UPI0001B7BC50 Ankyrin n=1 Tax=Rattus norvegicus RepID=... 73 1e-11
UniRef100_UPI0001B7BC48 Ankyrin n=1 Tax=Rattus norvegicus RepID=... 73 1e-11
UniRef100_Q8C8R3-5 Isoform 5 of Ankyrin-2 n=1 Tax=Mus musculus R... 73 1e-11
UniRef100_Q8C8R3-2 Isoform 2 of Ankyrin-2 n=2 Tax=Mus musculus R... 73 1e-11
UniRef100_Q8C8R3-4 Isoform 4 of Ankyrin-2 n=3 Tax=Mus musculus R... 73 1e-11
UniRef100_Q8C8R3 Ankyrin-2 n=1 Tax=Mus musculus RepID=ANK2_MOUSE 73 1e-11
UniRef100_UPI00017C322C PREDICTED: similar to ankyrin 2 n=1 Tax=... 72 2e-11
UniRef100_UPI0000E2053B PREDICTED: ankyrin 2 n=1 Tax=Pan troglod... 72 2e-11
UniRef100_UPI0000D9B200 PREDICTED: ankyrin 2, neuronal isoform 1... 72 2e-11
UniRef100_UPI0000D9B1FF PREDICTED: ankyrin 2, neuronal isoform 2... 72 2e-11
UniRef100_UPI0000D9B1FE PREDICTED: ankyrin 2 isoform 10 n=1 Tax=... 72 2e-11
UniRef100_UPI0000D9B1FD PREDICTED: ankyrin 2, neuronal isoform 8... 72 2e-11
UniRef100_UPI0000D9B1FC PREDICTED: ankyrin 2, neuronal isoform 6... 72 2e-11
UniRef100_UPI0000D9B1FB PREDICTED: ankyrin 2, neuronal isoform 4... 72 2e-11
UniRef100_UPI0000D9B1FA PREDICTED: ankyrin 2, neuronal isoform 7... 72 2e-11
UniRef100_UPI0000D9B1F9 PREDICTED: ankyrin 2, neuronal isoform 5... 72 2e-11
UniRef100_UPI0000D9B1F8 PREDICTED: ankyrin 2 isoform 13 n=1 Tax=... 72 2e-11
UniRef100_UPI0000D9B1F7 PREDICTED: ankyrin 2, neuronal isoform 1... 72 2e-11
UniRef100_UPI0000D9B1F6 PREDICTED: ankyrin 2 isoform 9 n=1 Tax=M... 72 2e-11
UniRef100_UPI0000D9B1F5 PREDICTED: ankyrin 2 isoform 11 n=1 Tax=... 72 2e-11
UniRef100_UPI0000D9B1F4 PREDICTED: ankyrin 2, neuronal isoform 3... 72 2e-11
UniRef100_UPI00005A54F7 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F6 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F5 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F4 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F3 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F2 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F1 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54F0 PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54EF PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54EE PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI00005A54ED PREDICTED: similar to ankyrin 2 isoform ... 72 2e-11
UniRef100_UPI0001AE7534 UPI0001AE7534 related cluster n=1 Tax=Ho... 72 2e-11
UniRef100_UPI0000EB045E Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (A... 72 2e-11
UniRef100_UPI000179ED09 PREDICTED: Bos taurus similar to ankyrin... 72 2e-11
UniRef100_A3FK32 Ankyrin repeat protein n=1 Tax=Oncopeltus fasci... 72 2e-11
UniRef100_B7Z651 cDNA FLJ61595, highly similar to Ankyrin-2 (Fra... 72 2e-11
UniRef100_B7Z636 cDNA FLJ55887, highly similar to Ankyrin-2 (Fra... 72 2e-11
UniRef100_A8MQV1 Putative uncharacterized protein ANK2 (Fragment... 72 2e-11
UniRef100_Q01484-5 Isoform 4 of Ankyrin-2 n=1 Tax=Homo sapiens R... 72 2e-11
UniRef100_Q01484-2 Isoform 2 of Ankyrin-2 n=1 Tax=Homo sapiens R... 72 2e-11
UniRef100_Q01484-4 Isoform 3 of Ankyrin-2 n=1 Tax=Homo sapiens R... 72 2e-11
UniRef100_Q01484 Ankyrin-2 n=2 Tax=Homo sapiens RepID=ANK2_HUMAN 72 2e-11
UniRef100_UPI000175F1BB PREDICTED: similar to ankyrin 2 n=1 Tax=... 71 4e-11
UniRef100_UPI0000D8CEFC UPI0000D8CEFC related cluster n=1 Tax=Da... 71 4e-11
UniRef100_UPI0000E49CF5 PREDICTED: similar to ankyrin 2,3/unc44 ... 69 1e-10
UniRef100_UPI0000E46E28 PREDICTED: similar to ankyrin 2,3/unc44 ... 69 1e-10
UniRef100_UPI00016E23AA UPI00016E23AA related cluster n=1 Tax=Ta... 69 1e-10
UniRef100_UPI00016E2390 UPI00016E2390 related cluster n=1 Tax=Ta... 69 1e-10
UniRef100_UPI00016E238F UPI00016E238F related cluster n=1 Tax=Ta... 69 1e-10
UniRef100_UPI00016E238E UPI00016E238E related cluster n=1 Tax=Ta... 69 1e-10
UniRef100_UPI00016E238D UPI00016E238D related cluster n=1 Tax=Ta... 69 1e-10
UniRef100_UPI0001797E8D PREDICTED: ankyrin 2, neuronal n=1 Tax=E... 69 2e-10
UniRef100_UPI00017B1F1D UPI00017B1F1D related cluster n=1 Tax=Te... 69 2e-10
UniRef100_UPI00017B1F1C UPI00017B1F1C related cluster n=1 Tax=Te... 69 2e-10
UniRef100_UPI00017B1F1B UPI00017B1F1B related cluster n=1 Tax=Te... 69 2e-10
UniRef100_Q4RKA6 Chromosome 18 SCAF15030, whole genome shotgun s... 69 2e-10
UniRef100_B5WW47 Ankyrin n=1 Tax=Burkholderia sp. H160 RepID=B5W... 69 2e-10
UniRef100_UPI0000E4A105 PREDICTED: similar to ankyrin 2,3/unc44 ... 69 3e-10
UniRef100_UPI00017B56B7 UPI00017B56B7 related cluster n=1 Tax=Te... 69 3e-10
UniRef100_Q4T089 Chromosome undetermined SCAF11289, whole genome... 69 3e-10
UniRef100_A2DUC8 Putative uncharacterized protein n=1 Tax=Tricho... 69 3e-10
UniRef100_UPI0000E49E22 PREDICTED: similar to GAC-1 n=1 Tax=Stro... 68 3e-10
UniRef100_UPI0000E47184 PREDICTED: similar to GAC-1 n=1 Tax=Stro... 68 3e-10
UniRef100_UPI0000E46C35 PREDICTED: similar to ankyrin 2,3/unc44 ... 68 3e-10
UniRef100_A2E1I7 Ankyrin repeat protein, putative n=1 Tax=Tricho... 68 3e-10
UniRef100_UPI000011264A UPI000011264A related cluster n=1 Tax=un... 68 4e-10
UniRef100_UPI0000E4A3F7 PREDICTED: similar to ankyrin 2,3/unc44 ... 68 4e-10
UniRef100_UPI0000E487B3 PREDICTED: similar to ankyrin 2,3/unc44 ... 68 4e-10
UniRef100_C3NJK3 Serine/threonine protein kinase n=1 Tax=Sulfolo... 68 4e-10
UniRef100_UPI0000E49045 PREDICTED: similar to ankyrin 2,3/unc44,... 67 6e-10
UniRef100_UPI0000E48260 PREDICTED: similar to ankyrin 2,3/unc44,... 67 6e-10
UniRef100_UPI00017B3B4F UPI00017B3B4F related cluster n=1 Tax=Te... 67 6e-10
UniRef100_UPI00016E3AF9 UPI00016E3AF9 related cluster n=1 Tax=Ta... 67 6e-10
UniRef100_UPI00016E3AF8 UPI00016E3AF8 related cluster n=1 Tax=Ta... 67 6e-10
UniRef100_UPI00016E3ADF UPI00016E3ADF related cluster n=1 Tax=Ta... 67 6e-10
UniRef100_UPI00016E3ADE UPI00016E3ADE related cluster n=1 Tax=Ta... 67 6e-10
UniRef100_Q4RYJ3 Chromosome 2 SCAF14976, whole genome shotgun se... 67 6e-10
UniRef100_A4RS51 Predicted protein n=1 Tax=Ostreococcus lucimari... 67 6e-10
UniRef100_A2FZU9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 67 6e-10
UniRef100_UPI000180D0A5 PREDICTED: similar to ankyrin 2 n=1 Tax=... 67 7e-10
UniRef100_UPI0000E4A75D PREDICTED: similar to ankyrin 2,3/unc44 ... 67 7e-10
UniRef100_UPI0000E47BA0 PREDICTED: similar to ankyrin 2,3/unc44 ... 67 7e-10
UniRef100_Q10AH9 Ankyrin repeat domain protein 2, putative, expr... 67 7e-10
UniRef100_Q851Y1 Os03g0851700 protein n=2 Tax=Oryza sativa RepID... 67 7e-10
UniRef100_UPI0000E48769 PREDICTED: similar to ankyrin 2,3/unc44,... 67 1e-09
UniRef100_B3EU24 Putative uncharacterized protein n=1 Tax=Candid... 67 1e-09
UniRef100_A2E6X8 Ankyrin repeat protein, putative n=1 Tax=Tricho... 67 1e-09
UniRef100_Q9SAR5 Ankyrin repeat domain-containing protein 2 n=3 ... 67 1e-09
UniRef100_UPI00001133CE UPI00001133CE related cluster n=1 Tax=un... 66 1e-09
UniRef100_C1BWB3 Cyclin-dependent kinase 4 inhibitor B n=1 Tax=E... 66 1e-09
UniRef100_UPI0000E4A970 PREDICTED: similar to ankyrin 2,3/unc44 ... 66 2e-09
UniRef100_UPI0000E47480 PREDICTED: similar to ankyrin 2,3/unc44 ... 66 2e-09
UniRef100_UPI0000E464A0 PREDICTED: similar to ankyrin 2,3/unc44 ... 66 2e-09
UniRef100_UPI0000E45FC1 PREDICTED: similar to ankyrin 2,3/unc44 ... 66 2e-09
UniRef100_UPI0001A2C309 UPI0001A2C309 related cluster n=1 Tax=Da... 66 2e-09
UniRef100_B5DE67 Putative uncharacterized protein (Fragment) n=1... 66 2e-09
UniRef100_A2G787 Ankyrin repeat protein, putative n=1 Tax=Tricho... 66 2e-09
UniRef100_A2DJE5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 66 2e-09
UniRef100_UPI0000E45D67 PREDICTED: similar to ankyrin 2,3/unc44 ... 65 2e-09
UniRef100_UPI0000DB765A PREDICTED: similar to ankyrin repeat dom... 65 2e-09
UniRef100_A7IUE3 Putative uncharacterized protein M413L n=1 Tax=... 65 2e-09
UniRef100_C6XC80 Ankyrin n=1 Tax=Methylovorus sp. SIP3-4 RepID=C... 65 2e-09
UniRef100_B1HW28 Putative ankyrin repeat protein n=1 Tax=Lysinib... 65 2e-09
UniRef100_A2E1J1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 65 2e-09
UniRef100_UPI0000E8257B PREDICTED: similar to ankyrin 3 n=1 Tax=... 65 3e-09
UniRef100_UPI0000E483A4 PREDICTED: similar to ankyrin 2,3/unc44 ... 65 3e-09
UniRef100_UPI0000589309 PREDICTED: similar to ankyrin 2,3/unc44 ... 65 3e-09
UniRef100_UPI0000ECB6A5 Ankyrin 3 n=1 Tax=Gallus gallus RepID=UP... 65 3e-09
UniRef100_UPI0000ECB6A4 Ankyrin 3 n=1 Tax=Gallus gallus RepID=UP... 65 3e-09
UniRef100_UPI0000ECB6A3 Ankyrin 3 n=1 Tax=Gallus gallus RepID=UP... 65 3e-09
UniRef100_UPI0000ECB68E Ankyrin 3 n=1 Tax=Gallus gallus RepID=UP... 65 3e-09
UniRef100_UPI0000ECB68D Ankyrin 3 n=1 Tax=Gallus gallus RepID=UP... 65 3e-09
UniRef100_A2BIB9 Novel protein similar to vertebrate ankyrin 2, ... 65 3e-09
UniRef100_A2E0J0 Putative uncharacterized protein n=1 Tax=Tricho... 65 3e-09
UniRef100_UPI0000E48FAC PREDICTED: similar to ankyrin 2,3/unc44 ... 65 4e-09
UniRef100_UPI0000DAED50 hypothetical protein Wendoof_01000169 n=... 65 4e-09
UniRef100_Q1H033 Ankyrin n=1 Tax=Methylobacillus flagellatus KT ... 65 4e-09
UniRef100_Q29Q26 At2g17390 n=1 Tax=Arabidopsis thaliana RepID=Q2... 65 4e-09
UniRef100_A2FAX8 Ankyrin repeat protein, putative n=1 Tax=Tricho... 65 4e-09
UniRef100_A2EG05 Ankyrin repeat protein, putative n=1 Tax=Tricho... 65 4e-09
UniRef100_UPI0000E48534 PREDICTED: similar to ankyrin 2,3/unc44 ... 64 5e-09
UniRef100_UPI0000E45CF7 PREDICTED: similar to ankyrin 2,3/unc44 ... 64 5e-09
UniRef100_B0SFK5 Ankyrin-repeat protein n=2 Tax=Leptospira bifle... 64 5e-09
UniRef100_A3I7E2 Ankyrin repeat domain protein n=1 Tax=Bacillus ... 64 5e-09
UniRef100_A2DXB5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 5e-09
UniRef100_A2DQT2 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 5e-09
UniRef100_A2DIK0 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 5e-09
UniRef100_A2D9Z2 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 5e-09
UniRef100_A2D9E7 Putative uncharacterized protein n=1 Tax=Tricho... 64 5e-09
UniRef100_UPI000194BC8B PREDICTED: ankyrin repeat domain 12 n=1 ... 64 6e-09
UniRef100_UPI0000E48D7C PREDICTED: similar to ankyrin 2,3/unc44,... 64 6e-09
UniRef100_UPI0000E47F89 PREDICTED: similar to ankyrin 2,3/unc44,... 64 6e-09
UniRef100_UPI0000E47F24 PREDICTED: similar to ankyrin 2,3/unc44 ... 64 6e-09
UniRef100_C1BWB7 Cyclin-dependent kinase 4 inhibitor B n=1 Tax=E... 64 6e-09
UniRef100_B7E764 cDNA clone:001-044-F07, full insert sequence n=... 64 6e-09
UniRef100_Q0J0F4 Os09g0513000 protein (Fragment) n=3 Tax=Oryza s... 64 6e-09
UniRef100_A2Z4X9 Putative uncharacterized protein n=1 Tax=Oryza ... 64 6e-09
UniRef100_Q16FI9 Ankyrin 2,3/unc44 (Fragment) n=1 Tax=Aedes aegy... 64 6e-09
UniRef100_A2ENB3 Putative uncharacterized protein n=1 Tax=Tricho... 64 6e-09
UniRef100_Q99PE2 Ankyrin repeat family A protein 2 n=1 Tax=Mus m... 64 6e-09
UniRef100_UPI0001554AD7 PREDICTED: similar to ankyrin repeat-con... 64 8e-09
UniRef100_UPI0001554AD6 PREDICTED: similar to ankyrin repeat-con... 64 8e-09
UniRef100_UPI0000F2B937 PREDICTED: similar to ankyrin repeat-con... 64 8e-09
UniRef100_UPI0000E7FEB8 PREDICTED: similar to GAC-1 n=1 Tax=Gall... 64 8e-09
UniRef100_UPI0000E45F8D PREDICTED: similar to ankyrin 2,3/unc44 ... 64 8e-09
UniRef100_Q5JSX5 Ankyrin 3, node of Ranvier (Ankyrin G) (Fragmen... 64 8e-09
UniRef100_UPI00016E2351 UPI00016E2351 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E2350 UPI00016E2350 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E2308 UPI00016E2308 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E2307 UPI00016E2307 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E2306 UPI00016E2306 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E22E7 UPI00016E22E7 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E22E6 UPI00016E22E6 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI00016E22E5 UPI00016E22E5 related cluster n=1 Tax=Ta... 64 8e-09
UniRef100_UPI0000ECCF29 Ankyrin repeat domain-containing protein... 64 8e-09
UniRef100_B5DQK5 GA23604 n=1 Tax=Drosophila pseudoobscura pseudo... 64 8e-09
UniRef100_B3RVS3 Putative uncharacterized protein n=1 Tax=Tricho... 64 8e-09
UniRef100_A2GGG6 Putative uncharacterized protein n=1 Tax=Tricho... 64 8e-09
UniRef100_A2EJ39 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 8e-09
UniRef100_A2D9U3 Ankyrin repeat protein, putative n=1 Tax=Tricho... 64 8e-09
UniRef100_Q0CV66 Putative uncharacterized protein n=1 Tax=Asperg... 64 8e-09
UniRef100_A1RZQ2 Ankyrin n=1 Tax=Thermofilum pendens Hrk 5 RepID... 64 8e-09
UniRef100_UPI0001795E2B PREDICTED: ankyrin repeat domain 11 n=1 ... 63 1e-08
UniRef100_UPI000176158B PREDICTED: similar to ankyrin 3 n=1 Tax=... 63 1e-08
UniRef100_UPI0000E4A7F8 PREDICTED: similar to ankyrin 2,3/unc44 ... 63 1e-08
UniRef100_UPI0000E48ACF PREDICTED: similar to ankyrin 2,3/unc44 ... 63 1e-08
UniRef100_UPI0000E47704 PREDICTED: similar to ankyrin 2,3/unc44 ... 63 1e-08
UniRef100_UPI0000E46EC5 PREDICTED: similar to ankyrin 2,3/unc44,... 63 1e-08
UniRef100_UPI0001A2DE8E UPI0001A2DE8E related cluster n=1 Tax=Da... 63 1e-08
UniRef100_UPI00001824C9 ankyrin repeat domain 39 n=1 Tax=Rattus ... 63 1e-08
UniRef100_UPI0000D61A35 Ankyrin repeat domain-containing protein... 63 1e-08
UniRef100_UPI00004569E1 ankyrin repeat domain 11 n=1 Tax=Homo sa... 63 1e-08
UniRef100_UPI000013E6C4 Ankyrin repeat domain-containing protein... 63 1e-08
UniRef100_UPI00016E8D56 UPI00016E8D56 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_Q49S16 Ankyrin domain protein (Fragment) n=1 Tax=Wolba... 63 1e-08
UniRef100_Q7XYD8 Apomixis-associated protein (Fragment) n=1 Tax=... 63 1e-08
UniRef100_A7PR02 Chromosome chr6 scaffold_25, whole genome shotg... 63 1e-08
UniRef100_A2GH03 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2FYB1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2FVV8 Ankyrin repeat protein, putative (Fragment) n=1... 63 1e-08
UniRef100_A2EY10 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2EFP9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2E159 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2E0C9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_Q9UHR3 Nasopharyngeal carcinoma susceptibility protein... 63 1e-08
UniRef100_Q59GC3 Ankyrin repeat domain 11 variant (Fragment) n=1... 63 1e-08
UniRef100_Q6UB99 Ankyrin repeat domain-containing protein 11 n=2... 63 1e-08
UniRef100_UPI0000E4A384 PREDICTED: similar to ankyrin 2,3/unc44,... 63 1e-08
UniRef100_UPI0000E4A383 PREDICTED: similar to ankyrin 2,3/unc44 ... 63 1e-08
UniRef100_UPI0000E477A4 PREDICTED: similar to ankyrin 2,3/unc44,... 63 1e-08
UniRef100_UPI0000E47748 PREDICTED: similar to ankyrin 2,3/unc44 ... 63 1e-08
UniRef100_UPI00005BEA8A PREDICTED: similar to ankyrin repeat dom... 63 1e-08
UniRef100_UPI000184A3E0 Ankyrin-3 (ANK-3) (Ankyrin-G). n=2 Tax=C... 63 1e-08
UniRef100_UPI00005A6055 PREDICTED: similar to ankyrin 3 isoform ... 63 1e-08
UniRef100_UPI00005A0D86 PREDICTED: similar to ankyrin repeat dom... 63 1e-08
UniRef100_UPI00005A06C9 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C8 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C7 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C6 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C5 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C4 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C3 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06C2 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI0000EB3E3D Ankyrin-3 (ANK-3) (Ankyrin-G). n=2 Tax=C... 63 1e-08
UniRef100_UPI00005A06C0 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06BF PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06BE PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06BD PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06BC PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06BB PREDICTED: similar to ankyrin 3 isoform ... 63 1e-08
UniRef100_UPI00005A06BA PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI0000EB3E3C Ankyrin-3 (ANK-3) (Ankyrin-G). n=2 Tax=C... 63 1e-08
UniRef100_UPI00005A06B7 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06B6 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_UPI00005A06B5 PREDICTED: similar to ankyrin 3, epithel... 63 1e-08
UniRef100_Q4RJD4 Chromosome 18 SCAF15038, whole genome shotgun s... 63 1e-08
UniRef100_UPI0001B79B00 UPI0001B79B00 related cluster n=1 Tax=Ra... 63 1e-08
UniRef100_UPI0001B79AFF UPI0001B79AFF related cluster n=1 Tax=Ra... 63 1e-08
UniRef100_UPI0000605E51 ankyrin repeat domain 11 n=1 Tax=Mus mus... 63 1e-08
UniRef100_UPI000024E220 ankyrin repeat domain 11 n=1 Tax=Mus mus... 63 1e-08
UniRef100_UPI00016E8D57 UPI00016E8D57 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_UPI00016E8D55 UPI00016E8D55 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_UPI000184A343 Ankyrin repeat domain-containing protein... 63 1e-08
UniRef100_UPI0000EB3E3E Ankyrin-3 (ANK-3) (Ankyrin-G). n=1 Tax=C... 63 1e-08
UniRef100_UPI0000EB3E1E Ankyrin-3 (ANK-3) (Ankyrin-G). n=1 Tax=C... 63 1e-08
UniRef100_UPI0000614A25 UPI0000614A25 related cluster n=1 Tax=Bo... 63 1e-08
UniRef100_Q9DES6 P13 n=1 Tax=Takifugu rubripes RepID=Q9DES6_TAKRU 63 1e-08
UniRef100_B5XAC9 26S proteasome non-ATPase regulatory subunit 10... 63 1e-08
UniRef100_Q05DP4 Ankrd11 protein (Fragment) n=1 Tax=Mus musculus... 63 1e-08
UniRef100_B7ZWM7 Ankrd11 protein n=1 Tax=Mus musculus RepID=B7ZW... 63 1e-08
UniRef100_C5HLB7 Ankyrin 2,3/unc44-like protein n=1 Tax=uncultur... 63 1e-08
UniRef100_Q7Q6V5 AGAP005648-PA n=1 Tax=Anopheles gambiae RepID=Q... 63 1e-08
UniRef100_A2G120 Ankyrin repeat protein, putative (Fragment) n=1... 63 1e-08
UniRef100_A2G119 Putative uncharacterized protein n=1 Tax=Tricho... 63 1e-08
UniRef100_A2G118 Putative uncharacterized protein n=1 Tax=Tricho... 63 1e-08
UniRef100_A2EK94 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2EDY5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 63 1e-08
UniRef100_A2DYP2 Putative uncharacterized protein n=1 Tax=Tricho... 63 1e-08
UniRef100_UPI000186F30E ankyrin repeat domain-containing protein... 62 2e-08
UniRef100_UPI000175FAED PREDICTED: similar to Ankyrin repeat dom... 62 2e-08
UniRef100_UPI000155F750 PREDICTED: similar to Ankyrin repeat, fa... 62 2e-08
UniRef100_UPI000155EB94 PREDICTED: ankyrin repeat domain 12 n=1 ... 62 2e-08
UniRef100_UPI0000E4A6BA PREDICTED: similar to ankyrin 2,3/unc44 ... 62 2e-08
UniRef100_UPI0000E48AD0 PREDICTED: similar to ankyrin 2,3/unc44 ... 62 2e-08
UniRef100_UPI0000E48535 PREDICTED: similar to ankyrin 2,3/unc44 ... 62 2e-08
UniRef100_UPI0000E24C64 PREDICTED: ankyrin repeat domain 12 isof... 62 2e-08
UniRef100_UPI0000E24C62 PREDICTED: ankyrin repeat domain 12 isof... 62 2e-08
UniRef100_UPI0000E24C61 PREDICTED: ankyrin repeat domain 12 isof... 62 2e-08
UniRef100_UPI0000E24469 PREDICTED: ankyrin repeat domain 11 n=1 ... 62 2e-08
UniRef100_UPI0000D554FF PREDICTED: similar to tankyrase n=1 Tax=... 62 2e-08
UniRef100_UPI00005BFD59 PREDICTED: similar to ankyrin repeat dom... 62 2e-08
UniRef100_UPI00004C0D1D PREDICTED: similar to ankyrin repeat dom... 62 2e-08
UniRef100_UPI00004292D2 PREDICTED: similar to Ankrd12 protein n=... 62 2e-08
UniRef100_UPI0000D8E40B UPI0000D8E40B related cluster n=1 Tax=Da... 62 2e-08
UniRef100_UPI000069E7FF Ankyrin repeat domain-containing protein... 62 2e-08
UniRef100_Q3U569 Putative uncharacterized protein (Fragment) n=2... 62 2e-08
UniRef100_UPI0000EB39B3 Ankyrin repeat domain-containing protein... 62 2e-08
UniRef100_UPI00004C0D1C PREDICTED: similar to ankyrin repeat dom... 62 2e-08
UniRef100_UPI00004A4027 PREDICTED: similar to ankyrin repeat, fa... 62 2e-08
UniRef100_UPI00005BFD58 UPI00005BFD58 related cluster n=1 Tax=Bo... 62 2e-08
UniRef100_Q9W6I8 P13CDKN2X n=1 Tax=Xiphophorus hellerii RepID=Q9... 62 2e-08
UniRef100_Q6GQ19 MGC80441 protein n=1 Tax=Xenopus laevis RepID=Q... 62 2e-08
UniRef100_P70067 13CDKN2X protein n=1 Tax=Xiphophorus maculatus ... 62 2e-08
UniRef100_Q80UL6 Ankrd12 protein (Fragment) n=1 Tax=Mus musculus... 62 2e-08
UniRef100_Q66JQ1 Ankrd12 protein (Fragment) n=1 Tax=Mus musculus... 62 2e-08
UniRef100_Q3UZR0 Putative uncharacterized protein n=1 Tax=Mus mu... 62 2e-08
UniRef100_Q05CM6 Ankrd12 protein (Fragment) n=1 Tax=Mus musculus... 62 2e-08
UniRef100_Q5NVJ2 Putative uncharacterized protein DKFZp459A1520 ... 62 2e-08
UniRef100_C4Q910 Ankyrin 2,3/unc44, putative n=1 Tax=Schistosoma... 62 2e-08
UniRef100_C4Q0F9 Tankyrase, putative n=1 Tax=Schistosoma mansoni... 62 2e-08
UniRef100_A2EWB1 Putative uncharacterized protein n=1 Tax=Tricho... 62 2e-08
UniRef100_A2DG32 Putative uncharacterized protein n=1 Tax=Tricho... 62 2e-08
UniRef100_Q6PG48 ANKRD12 protein n=1 Tax=Homo sapiens RepID=Q6PG... 62 2e-08
UniRef100_Q6NSE1 ANKRD12 protein (Fragment) n=1 Tax=Homo sapiens... 62 2e-08
UniRef100_Q2KI79 Ankyrin repeat family A protein 2 n=1 Tax=Bos t... 62 2e-08
UniRef100_Q6UB98-2 Isoform 2 of Ankyrin repeat domain-containing... 62 2e-08
UniRef100_Q6UB98 Ankyrin repeat domain-containing protein 12 n=1... 62 2e-08
UniRef100_UPI000186DD6F protein phosphatase 1 regulatory subunit... 62 2e-08
UniRef100_UPI0001797E80 PREDICTED: ankyrin 3, node of Ranvier (a... 62 2e-08
UniRef100_UPI0000E80DD0 PREDICTED: similar to ankyrin repeat-con... 62 2e-08
UniRef100_UPI0000E1F595 PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI00005C412D ankyrin 3, epithelial isoform 2 n=1 Tax=... 62 2e-08
UniRef100_UPI00005C412C ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0001B79ABC ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0001B79ABA ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0001B79AB9 ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0001B79AB7 ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0001B79AB6 ankyrin 3, epithelial isoform 1 n=1 Tax=... 62 2e-08
UniRef100_UPI0000DBF42B UPI0000DBF42B related cluster n=1 Tax=Ra... 62 2e-08
UniRef100_UPI0000DBF42A UPI0000DBF42A related cluster n=1 Tax=Ra... 62 2e-08
UniRef100_UPI00005075A7 ankyrin repeat domain 12 n=1 Tax=Rattus ... 62 2e-08
UniRef100_UPI00004A5279 PREDICTED: similar to ankyrin repeat dom... 62 2e-08
UniRef100_UPI0000F34A38 UPI0000F34A38 related cluster n=1 Tax=Bo... 62 2e-08
UniRef100_UPI0000ECAE9E Ankyrin repeat domain-containing protein... 62 2e-08
UniRef100_UPI0000ECAE9D Ankyrin repeat domain-containing protein... 62 2e-08
UniRef100_A2BIM2 Si:ch211-203b8.6 n=1 Tax=Danio rerio RepID=A2BI... 62 2e-08
UniRef100_Q574D8 Ankyrin G197 n=1 Tax=Rattus norvegicus RepID=Q5... 62 2e-08
UniRef100_Q574D7 Ankyrin G217 n=1 Tax=Rattus norvegicus RepID=Q5... 62 2e-08
UniRef100_O88521 190 kDa ankyrin isoform n=1 Tax=Rattus norvegic... 62 2e-08
UniRef100_O70511 270 kDa ankyrin G isoform (Fragment) n=1 Tax=Ra... 62 2e-08
UniRef100_B5UNN0 Ankyrin repeat domain protein n=1 Tax=Bacillus ... 62 2e-08
UniRef100_Q00S14 FOG: Ankyrin repeat (ISS) n=1 Tax=Ostreococcus ... 62 2e-08
UniRef100_B9S453 Ankyrin repeat-containing protein, putative n=1... 62 2e-08
UniRef100_C3Y2F3 Putative uncharacterized protein n=1 Tax=Branch... 62 2e-08
UniRef100_A2EU43 Putative uncharacterized protein n=1 Tax=Tricho... 62 2e-08
UniRef100_A2ELJ6 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2E9R6 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2E3W8 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2DS48 PH domain containing protein n=1 Tax=Trichomona... 62 2e-08
UniRef100_A2DRS8 Histone-lysine N-methyltransferase, H3 lysine-9... 62 2e-08
UniRef100_A2DJY8 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2DF79 Putative uncharacterized protein n=1 Tax=Tricho... 62 2e-08
UniRef100_A2DEH1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2DCA1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2D7K9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_A2D779 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 2e-08
UniRef100_B2R5H9 cDNA, FLJ92480 n=1 Tax=Homo sapiens RepID=B2R5H... 62 2e-08
UniRef100_Q53RE8 Ankyrin repeat domain-containing protein 39 n=1... 62 2e-08
UniRef100_UPI00017F055A PREDICTED: ankyrin 3, node of Ranvier (a... 62 3e-08
UniRef100_UPI0001792158 PREDICTED: similar to rolling pebbles n=... 62 3e-08
UniRef100_UPI0000E4A3F8 PREDICTED: similar to ankyrin 2,3/unc44 ... 62 3e-08
UniRef100_UPI0000E49DD6 PREDICTED: similar to ankyrin 2,3/unc44 ... 62 3e-08
UniRef100_UPI0000E49251 PREDICTED: similar to ankyrin 2,3/unc44,... 62 3e-08
UniRef100_UPI0000E487B2 PREDICTED: similar to ankyrin 2,3/unc44 ... 62 3e-08
UniRef100_UPI0000E4725D PREDICTED: similar to ankyrin 2,3/unc44 ... 62 3e-08
UniRef100_UPI0000D9B4C9 PREDICTED: similar to ankyrin repeat, fa... 62 3e-08
UniRef100_UPI0000D9B4C8 PREDICTED: similar to ankyrin repeat, fa... 62 3e-08
UniRef100_UPI000036D0D0 PREDICTED: hypothetical protein isoform ... 62 3e-08
UniRef100_UPI00017B58AE UPI00017B58AE related cluster n=1 Tax=Te... 62 3e-08
UniRef100_UPI00017B58AD UPI00017B58AD related cluster n=1 Tax=Te... 62 3e-08
UniRef100_UPI0001AE79C7 UPI0001AE79C7 related cluster n=1 Tax=Ho... 62 3e-08
UniRef100_UPI0001AE6D3E Ankyrin-3 (ANK-3) (Ankyrin-G). n=1 Tax=H... 62 3e-08
UniRef100_UPI0001AE6D3D Ankyrin-3 (ANK-3) (Ankyrin-G). n=1 Tax=H... 62 3e-08
UniRef100_UPI000049DCF2 Ankyrin-3 (ANK-3) (Ankyrin-G). n=1 Tax=H... 62 3e-08
UniRef100_Q4RAH4 Chromosome undetermined SCAF23648, whole genome... 62 3e-08
UniRef100_B3TLA9 Cell cycle inhibitor p15 n=1 Tax=Nothobranchius... 62 3e-08
UniRef100_A2BID2 Novel protein containing ankyrin repeats n=1 Ta... 62 3e-08
UniRef100_C0QTZ7 Pfs, nacht and ankyrin domain protein n=1 Tax=P... 62 3e-08
UniRef100_C3DRE7 Ankyrin repeat domain protein n=1 Tax=Bacillus ... 62 3e-08
UniRef100_C2NB00 Ankyrin repeat domain protein n=1 Tax=Bacillus ... 62 3e-08
UniRef100_A6EG62 Ankyrin-like protein n=1 Tax=Pedobacter sp. BAL... 62 3e-08
UniRef100_Q40785 Pennisetum ciliare possible apospory-associated... 62 3e-08
UniRef100_Q019U9 Ankyrin repeat protein E4_2 (ISS) n=1 Tax=Ostre... 62 3e-08
UniRef100_Q5RDK9 Putative uncharacterized protein DKFZp459D1861 ... 62 3e-08
UniRef100_A7SZG9 Predicted protein n=1 Tax=Nematostella vectensi... 62 3e-08
UniRef100_A2FAC4 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 3e-08
UniRef100_A2F9F9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 3e-08
UniRef100_A2E7A5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 62 3e-08
UniRef100_A2DAB3 Putative uncharacterized protein n=1 Tax=Tricho... 62 3e-08
UniRef100_Q7Z3G4 Putative uncharacterized protein DKFZp686P17114... 62 3e-08
UniRef100_Q5CZH9 Putative uncharacterized protein DKFZp686I225 n... 62 3e-08
UniRef100_Q59G01 Ankyrin 3 isoform 1 variant (Fragment) n=1 Tax=... 62 3e-08
UniRef100_Q13484 Ankyrin G119 n=1 Tax=Homo sapiens RepID=Q13484_... 62 3e-08
UniRef100_B4DIL1 cDNA FLJ58990, highly similar to Ankyrin-3 n=1 ... 62 3e-08
UniRef100_B6QSL1 Ankyrin repeat-containing protein, putative n=1... 62 3e-08
UniRef100_Q9H9E1 Ankyrin repeat family A protein 2 n=1 Tax=Homo ... 62 3e-08
UniRef100_Q12955 Ankyrin-3 n=1 Tax=Homo sapiens RepID=ANK3_HUMAN 62 3e-08
UniRef100_UPI00015B54EB PREDICTED: similar to protein phosphatas... 61 4e-08
UniRef100_UPI0000F2EC84 PREDICTED: similar to ankyrin 3 n=1 Tax=... 61 4e-08
UniRef100_UPI0000E4A9B6 PREDICTED: similar to ankyrin 2,3/unc44 ... 61 4e-08
UniRef100_UPI0000E46EFE PREDICTED: similar to ankyrin 2,3/unc44 ... 61 4e-08
UniRef100_UPI000051AA30 PREDICTED: similar to MYPT-75D CG6896-PA... 61 4e-08
UniRef100_UPI00004D4204 Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tanky... 61 4e-08
UniRef100_UPI00017B130B UPI00017B130B related cluster n=1 Tax=Te... 61 4e-08
UniRef100_UPI00017B130A UPI00017B130A related cluster n=1 Tax=Te... 61 4e-08
UniRef100_UPI00016E260E UPI00016E260E related cluster n=1 Tax=Ta... 61 4e-08
UniRef100_UPI00016E260D UPI00016E260D related cluster n=1 Tax=Ta... 61 4e-08
UniRef100_UPI00016E260C UPI00016E260C related cluster n=1 Tax=Ta... 61 4e-08
UniRef100_Q5XGK5 LOC495279 protein n=1 Tax=Xenopus laevis RepID=... 61 4e-08
UniRef100_Q4S372 Chromosome 4 SCAF14752, whole genome shotgun se... 61 4e-08
UniRef100_B3ETJ2 Putative uncharacterized protein n=1 Tax=Candid... 61 4e-08
UniRef100_A8PKG0 Ankrd17 protein n=1 Tax=Rickettsiella grylli Re... 61 4e-08
UniRef100_Q01FG9 Ankyrin repeat family protein / AFT protein (IS... 61 4e-08
UniRef100_C5WS25 Putative uncharacterized protein Sb01g000990 n=... 61 4e-08
UniRef100_B4FRE3 Ankyrin repeat domain-containing protein 2 n=1 ... 61 4e-08
UniRef100_Q16FJ0 Serine/threonine-protein kinase ripk4 (Fragment... 61 4e-08
UniRef100_A2GYW5 Putative uncharacterized protein (Fragment) n=1... 61 4e-08
UniRef100_A2G8M9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 4e-08
UniRef100_A2FME9 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 4e-08
UniRef100_A2F2X3 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 4e-08
UniRef100_A2ENN0 Espin, putative n=1 Tax=Trichomonas vaginalis G... 61 4e-08
UniRef100_A2EKK2 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 4e-08
UniRef100_A2EF48 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 4e-08
UniRef100_UPI000194E1AE PREDICTED: ankyrin repeat, family A (RFX... 61 5e-08
UniRef100_UPI0001925AC2 PREDICTED: similar to predicted protein,... 61 5e-08
UniRef100_UPI00017970A5 PREDICTED: similar to ankyrin repeat dom... 61 5e-08
UniRef100_UPI0000E493E6 PREDICTED: similar to ankyrin 2,3/unc44 ... 61 5e-08
UniRef100_UPI0000E492FD PREDICTED: similar to ankyrin 2,3/unc44 ... 61 5e-08
UniRef100_UPI0000E48ACE PREDICTED: similar to ankyrin 2,3/unc44 ... 61 5e-08
UniRef100_UPI0000E47718 PREDICTED: similar to ankyrin 2,3/unc44 ... 61 5e-08
UniRef100_UPI0000E45C19 PREDICTED: similar to ankyrin 2,3/unc44 ... 61 5e-08
UniRef100_UPI0000588EF6 PREDICTED: hypothetical protein n=1 Tax=... 61 5e-08
UniRef100_UPI00016E958E UPI00016E958E related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E94AD UPI00016E94AD related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E94AC UPI00016E94AC related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E94AB UPI00016E94AB related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E94AA UPI00016E94AA related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E1164 UPI00016E1164 related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E1146 UPI00016E1146 related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI00016E1145 UPI00016E1145 related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_Q6TGV5 Proteasome 26S subunit, non-ATPase, 10 n=1 Tax=... 61 5e-08
UniRef100_Q5ZIC2 Putative uncharacterized protein n=1 Tax=Gallus... 61 5e-08
UniRef100_Q4VBW1 Proteasome (Prosome, macropain) 26S subunit, no... 61 5e-08
UniRef100_C6ERD7 Cell cycle inhibitor p15 n=1 Tax=Nothobranchius... 61 5e-08
UniRef100_C1BJE4 26S proteasome non-ATPase regulatory subunit 10... 61 5e-08
UniRef100_B1ZVE9 Putative uncharacterized protein n=1 Tax=Opitut... 61 5e-08
UniRef100_Q69U94 Putative ankyrin domain protein n=1 Tax=Oryza s... 61 5e-08
UniRef100_C6T063 Putative uncharacterized protein n=1 Tax=Glycin... 61 5e-08
UniRef100_B9GZ26 Predicted protein n=1 Tax=Populus trichocarpa R... 61 5e-08
UniRef100_B9G212 Putative uncharacterized protein n=1 Tax=Oryza ... 61 5e-08
UniRef100_A9RGG9 Predicted protein n=1 Tax=Physcomitrella patens... 61 5e-08
UniRef100_A2G715 Inversin protein alternative isoform, putative ... 61 5e-08
UniRef100_A2FG57 Putative uncharacterized protein n=1 Tax=Tricho... 61 5e-08
UniRef100_A2F7J5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2F776 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2F767 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2EZL4 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2ENW5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2EE21 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2E8W4 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2DQU7 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2DFS5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_A2DBS1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 61 5e-08
UniRef100_Q9J4Z6 Putative ankyrin repeat protein FPV244 n=1 Tax=... 61 5e-08
UniRef100_UPI0001757F7D PREDICTED: similar to ankyrin 2,3/unc44 ... 60 7e-08
UniRef100_UPI0000E4A708 PREDICTED: similar to ankyrin 2,3/unc44 ... 60 7e-08
UniRef100_UPI0000E49CE7 PREDICTED: similar to ankyrin 2,3/unc44,... 60 7e-08
UniRef100_UPI0000E470F4 PREDICTED: similar to ankyrin 2,3/unc44,... 60 7e-08
UniRef100_UPI0000E21D24 PREDICTED: similar to mFLJ00392 protein ... 60 7e-08
UniRef100_UPI0000D9C13E PREDICTED: similar to protein phosphatas... 60 7e-08
UniRef100_UPI00006D3C2F PREDICTED: similar to protein phosphatas... 60 7e-08
UniRef100_UPI000058466F PREDICTED: similar to ankyrin 2,3/unc44 ... 60 7e-08
UniRef100_Q9XZ37 Putative uncharacterized protein n=1 Tax=Drosop... 60 7e-08
UniRef100_Q9VBP3 FI03751p n=1 Tax=Drosophila melanogaster RepID=... 60 7e-08
UniRef100_Q295L9 GA18382 n=1 Tax=Drosophila pseudoobscura pseudo... 60 7e-08
UniRef100_B4QVC8 GD18183 n=1 Tax=Drosophila simulans RepID=B4QVC... 60 7e-08
UniRef100_B4PTI2 GE10679 n=1 Tax=Drosophila yakuba RepID=B4PTI2_... 60 7e-08
UniRef100_B4K4W1 GI10353 n=1 Tax=Drosophila mojavensis RepID=B4K... 60 7e-08
UniRef100_B4JHT7 GH18604 n=1 Tax=Drosophila grimshawi RepID=B4JH... 60 7e-08
UniRef100_B4ICC0 GM10230 n=1 Tax=Drosophila sechellia RepID=B4IC... 60 7e-08
UniRef100_B4GG04 GL22305 n=1 Tax=Drosophila persimilis RepID=B4G... 60 7e-08
UniRef100_B3P6G9 GG12231 n=1 Tax=Drosophila erecta RepID=B3P6G9_... 60 7e-08
UniRef100_B3M617 GF23770 n=1 Tax=Drosophila ananassae RepID=B3M6... 60 7e-08
UniRef100_B3LYN0 GF18240 n=1 Tax=Drosophila ananassae RepID=B3LY... 60 7e-08
UniRef100_A2GMJ5 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_A2G5Q1 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_A2FS53 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_A2F7B7 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_A2DU09 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_A2D927 Ankyrin repeat protein, putative n=1 Tax=Tricho... 60 7e-08
UniRef100_C9SB95 Ankyrin-1 n=1 Tax=Verticillium albo-atrum VaMs.... 60 7e-08
UniRef100_B8N300 Ankyrin repeat-containing protein, putative n=1... 60 7e-08
UniRef100_Q96I34 Protein phosphatase 1 regulatory subunit 16A n=... 60 7e-08
UniRef100_UPI000194BC8F PREDICTED: oxysterol binding protein-lik... 60 9e-08