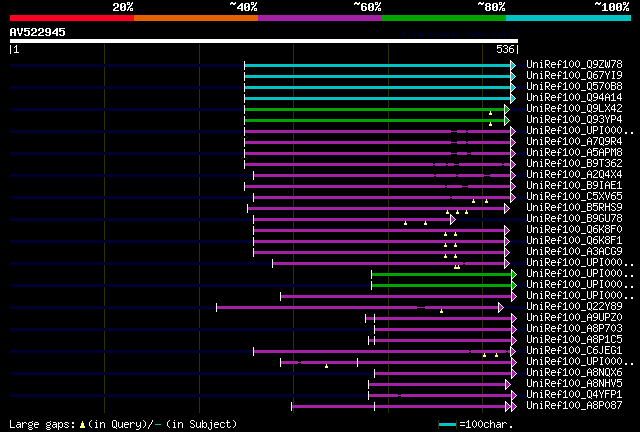
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV522945 APZL10g01F
(536 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q9ZW78 Putative uncharacterized protein At2g43170 n=1 ... 207 3e-52
UniRef100_Q67YI9 Putative uncharacterized protein At2g43170 n=1 ... 207 3e-52
UniRef100_Q570B8 Putative uncharacterized protein At2g43170 n=1 ... 207 3e-52
UniRef100_Q94A14 At2g43170/F14B2.11 n=1 Tax=Arabidopsis thaliana... 205 2e-51
UniRef100_Q9LX42 Epsin-like protein n=1 Tax=Arabidopsis thaliana... 117 4e-25
UniRef100_Q93YP4 Epsin-like protein n=1 Tax=Arabidopsis thaliana... 117 4e-25
UniRef100_UPI0001983401 PREDICTED: hypothetical protein n=1 Tax=... 86 2e-15
UniRef100_A7Q9R4 Chromosome chr5 scaffold_67, whole genome shotg... 86 2e-15
UniRef100_A5APM8 Putative uncharacterized protein n=1 Tax=Vitis ... 85 4e-15
UniRef100_B9T362 Epsin-2, putative n=1 Tax=Ricinus communis RepI... 80 1e-13
UniRef100_A2Q4X4 Epsin, N-terminal; ENTH/VHS n=1 Tax=Medicago tr... 75 2e-12
UniRef100_B9IAE1 Predicted protein (Fragment) n=1 Tax=Populus tr... 75 4e-12
UniRef100_C5XV65 Putative uncharacterized protein Sb04g036550 n=... 73 1e-11
UniRef100_B5RHS9 Epsin N-terminal homology (ENTH) domain-contain... 72 3e-11
UniRef100_B9GU78 Predicted protein n=1 Tax=Populus trichocarpa R... 69 2e-10
UniRef100_Q6K8F0 Epsin-like n=1 Tax=Oryza sativa Japonica Group ... 68 5e-10
UniRef100_Q6K8F1 Os02g0806600 protein n=3 Tax=Oryza sativa RepID... 68 5e-10
UniRef100_A3ACG9 Putative uncharacterized protein n=1 Tax=Oryza ... 66 1e-09
UniRef100_UPI000187CAA6 hypothetical protein MPER_03762 n=1 Tax=... 57 6e-07
UniRef100_UPI00006A15FE UPI00006A15FE related cluster n=1 Tax=Xe... 57 1e-06
UniRef100_UPI00006A15FD UPI00006A15FD related cluster n=1 Tax=Xe... 57 1e-06
UniRef100_UPI0000DBF811 UPI0000DBF811 related cluster n=1 Tax=Ra... 56 2e-06
UniRef100_Q22Y89 GAT domain containing protein n=1 Tax=Tetrahyme... 56 2e-06
UniRef100_A9UPZ0 Predicted protein n=1 Tax=Monosiga brevicollis ... 56 2e-06
UniRef100_A8P703 Histidine-rich glycoprotein, putative n=1 Tax=B... 56 2e-06
UniRef100_A8P1C5 T-cell receptor beta chain ANA 11, putative (Fr... 56 2e-06
UniRef100_C6JEG1 Putative uncharacterized protein n=1 Tax=Rumino... 55 2e-06
UniRef100_UPI0000DC0177 UPI0000DC0177 related cluster n=1 Tax=Ra... 55 3e-06
UniRef100_A8NQX6 T-cell receptor beta chain ANA 11, putative n=1... 55 4e-06
UniRef100_A8NHV5 T-cell receptor beta chain ANA 11, putative (Fr... 55 4e-06
UniRef100_Q4YFP1 Putative uncharacterized protein (Fragment) n=1... 54 5e-06
UniRef100_A8P087 T-cell receptor beta chain ANA 11, putative (Fr... 54 7e-06
UniRef100_UPI0000DC0B84 UPI0000DC0B84 related cluster n=1 Tax=Ra... 54 9e-06