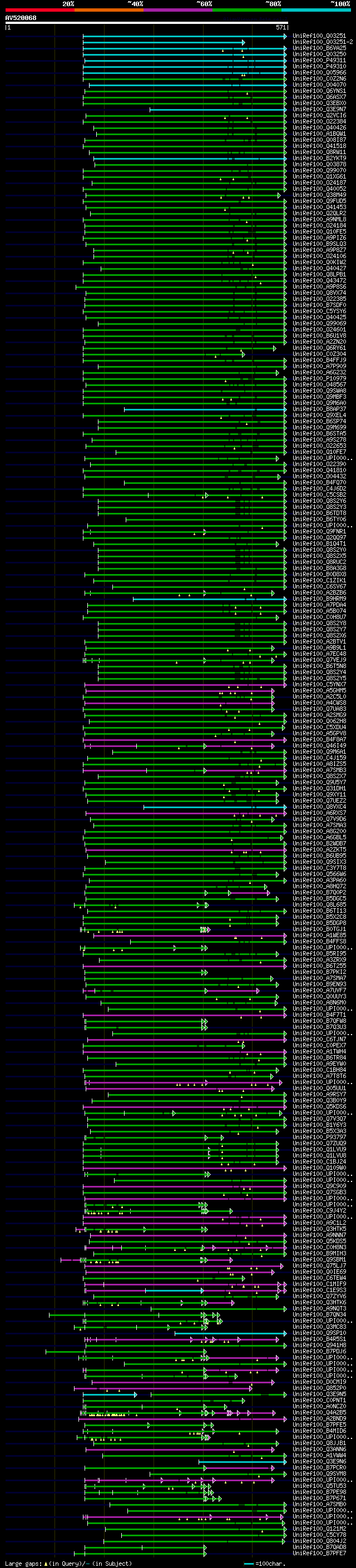
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV520068 APZ05h11F
(571 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q03251 Glycine-rich RNA-binding protein 8 n=2 Tax=Arab... 297 3e-79
UniRef100_Q03251-2 Isoform 2 of Glycine-rich RNA-binding protein... 243 7e-63
UniRef100_B6VA25 Putative glycine-rich RNA-binding protein n=1 T... 229 1e-58
UniRef100_Q03250 Glycine-rich RNA-binding protein 7 n=1 Tax=Arab... 227 6e-58
UniRef100_P49311 Glycine-rich RNA-binding protein GRP2A n=1 Tax=... 222 1e-56
UniRef100_P49310 Glycine-rich RNA-binding protein GRP1A n=1 Tax=... 218 3e-55
UniRef100_Q05966 Glycine-rich RNA-binding protein 10 n=1 Tax=Bra... 212 2e-53
UniRef100_C0Z2N6 AT2G21660 protein n=1 Tax=Arabidopsis thaliana ... 198 3e-49
UniRef100_O04070 SGRP-1 protein n=1 Tax=Solanum commersonii RepI... 197 5e-49
UniRef100_Q6YNS1 Putative glycine-rich RNA-binding protein n=1 T... 196 1e-48
UniRef100_Q6ASX7 Glycine-rich RNA binding protein n=1 Tax=Oryza ... 196 1e-48
UniRef100_Q3EBX0 Putative uncharacterized protein At2g21660.2 n=... 196 1e-48
UniRef100_Q3E9N7 Putative uncharacterized protein At4g39260.2 n=... 194 3e-48
UniRef100_Q2VCI6 Putative glycine-rich RNA binding protein-like ... 192 1e-47
UniRef100_O22384 Glycine-rich protein n=1 Tax=Oryza sativa RepID... 192 1e-47
UniRef100_Q40426 RNA-binding glycine-rich protein-1a n=1 Tax=Nic... 190 6e-47
UniRef100_A1BQW1 Glycine-rich RNA-binding protein (Fragment) n=1... 189 1e-46
UniRef100_Q08I87 Putative glycine-rich RNA-binding protein n=1 T... 188 2e-46
UniRef100_Q41518 Single-stranded nucleic acid binding protein n=... 188 3e-46
UniRef100_Q8RW11 Putative glycine rich protein n=1 Tax=Rumex obt... 187 4e-46
UniRef100_B2YKT9 Glycine-rich RNA-binding protein n=1 Tax=Nicoti... 187 5e-46
UniRef100_Q03878 Glycine-rich RNA-binding protein n=1 Tax=Daucus... 187 5e-46
UniRef100_Q99070 Glycine-rich RNA-binding protein 2 n=1 Tax=Sorg... 186 1e-45
UniRef100_Q1XG61 Putative glycine-rich RNA binding protein n=1 T... 184 3e-45
UniRef100_O24187 OsGRP1 n=1 Tax=Oryza sativa RepID=O24187_ORYSA 184 4e-45
UniRef100_Q40052 Glycine rich protein, RNA binding protein n=1 T... 184 5e-45
UniRef100_Q38M49 Putative glycine-rich RNA binding protein-like ... 183 7e-45
UniRef100_Q9FUD5 Glycine-rich RNA-binding protein n=1 Tax=Sorghu... 182 1e-44
UniRef100_Q41453 Putative glycine rich RNA binding protein n=1 T... 182 1e-44
UniRef100_Q2QLR2 Os12g0632000 protein n=1 Tax=Oryza sativa Japon... 182 2e-44
UniRef100_A9NML8 Putative uncharacterized protein n=1 Tax=Picea ... 181 3e-44
UniRef100_O24184 Glycine-rich RNA-binding protein n=1 Tax=Oryza ... 181 3e-44
UniRef100_Q10FE5 Retrotransposon protein, putative, Ty1-copia su... 181 5e-44
UniRef100_A9PIZ6 Putative uncharacterized protein n=1 Tax=Populu... 180 8e-44
UniRef100_B9SLQ3 Glycine-rich RNA-binding protein, putative n=1 ... 179 1e-43
UniRef100_A9P8Z7 Predicted protein n=1 Tax=Populus trichocarpa R... 179 1e-43
UniRef100_O24106 RNA-binding protein n=1 Tax=Nicotiana glutinosa... 179 1e-43
UniRef100_Q0KIW2 Glycine-rich RNA-binding protein n=1 Tax=Tritic... 179 2e-43
UniRef100_Q40427 RNA-binding glycine-rich protein-1b n=1 Tax=Nic... 178 2e-43
UniRef100_Q8LPB1 Glycine-rich RNA-binding protein n=1 Tax=Physco... 178 3e-43
UniRef100_Q43472 Low temperature-responsive RNA-binding protein ... 177 4e-43
UniRef100_A9P8S6 Putative uncharacterized protein n=1 Tax=Populu... 177 4e-43
UniRef100_Q8VX74 Glycine-rich RNA-binding protein n=1 Tax=Ricinu... 177 5e-43
UniRef100_O22385 Glycine-rich protein n=1 Tax=Oryza sativa RepID... 177 5e-43
UniRef100_B7SDF0 Glycine-rich RNA binding protein n=1 Tax=Oryza ... 177 5e-43
UniRef100_C5YSY6 Putative uncharacterized protein Sb08g022740 n=... 176 1e-42
UniRef100_Q40425 RNA-binding gricine-rich protein-1c n=1 Tax=Nic... 175 2e-42
UniRef100_Q99069 Glycine-rich RNA-binding protein 1 (Fragment) n... 175 2e-42
UniRef100_O24601 Glycine-rich RNA binding protein 1 n=1 Tax=Pela... 175 3e-42
UniRef100_B6U1V8 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 175 3e-42
UniRef100_A2ZN20 Putative uncharacterized protein n=1 Tax=Oryza ... 175 3e-42
UniRef100_Q6RY61 Glycine-rich RNA-binding protein RGP-1c (Fragme... 174 6e-42
UniRef100_C0Z304 AT2G21660 protein n=2 Tax=Arabidopsis thaliana ... 174 6e-42
UniRef100_B4FFJ9 Putative uncharacterized protein n=1 Tax=Zea ma... 173 7e-42
UniRef100_A7P909 Chromosome chr3 scaffold_8, whole genome shotgu... 173 1e-41
UniRef100_A6G232 RNA-binding protein n=1 Tax=Plesiocystis pacifi... 172 2e-41
UniRef100_P10979 Glycine-rich RNA-binding, abscisic acid-inducib... 172 2e-41
UniRef100_O48567 Glycine-rich RNA-binding protein n=1 Tax=Euphor... 171 3e-41
UniRef100_Q9SWA8 Glycine-rich RNA-binding protein n=1 Tax=Glycin... 171 5e-41
UniRef100_Q9MBF3 Glycine-rich RNA-binding protein n=1 Tax=Citrus... 168 3e-40
UniRef100_Q9M6A0 Putative glycine-rich RNA binding protein 3 n=1... 166 1e-39
UniRef100_B8AP37 Putative uncharacterized protein n=1 Tax=Oryza ... 165 2e-39
UniRef100_Q9XEL4 Glycine-rich RNA-binding protein n=1 Tax=Picea ... 163 1e-38
UniRef100_B6SP74 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 163 1e-38
UniRef100_Q9M699 Putative glycine-rich RNA-binding protein 2 n=1... 160 6e-38
UniRef100_B6STA5 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 160 8e-38
UniRef100_A9S278 Predicted protein n=2 Tax=Physcomitrella patens... 159 1e-37
UniRef100_O22653 Glycine-rich RNA-binding protein n=1 Tax=Euphor... 158 2e-37
UniRef100_Q10FE7 Os03g0670700 protein n=1 Tax=Oryza sativa Japon... 158 3e-37
UniRef100_UPI000180C465 PREDICTED: similar to cold-inducible RNA... 157 4e-37
UniRef100_O22390 Glycine-rich protein n=1 Tax=Oryza sativa RepID... 156 9e-37
UniRef100_Q41810 Glycine-rich protein n=1 Tax=Zea mays RepID=Q41... 156 1e-36
UniRef100_O04432 Glycine-rich protein n=1 Tax=Oryza sativa RepID... 155 2e-36
UniRef100_B4FQ70 Putative uncharacterized protein n=1 Tax=Zea ma... 155 3e-36
UniRef100_C4J6D2 Putative uncharacterized protein n=1 Tax=Zea ma... 153 8e-36
UniRef100_C5CSB2 RNP-1 like RNA-binding protein n=1 Tax=Variovor... 152 1e-35
UniRef100_Q8S2Y6 Glycine-rich RNA binding protein (Fragment) n=3... 152 1e-35
UniRef100_Q8S2Y3 Glycine-rich RNA binding protein (Fragment) n=2... 152 1e-35
UniRef100_B6TDT8 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 152 1e-35
UniRef100_B6TY06 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 152 2e-35
UniRef100_UPI0001862007 hypothetical protein BRAFLDRAFT_58365 n=... 151 4e-35
UniRef100_Q9FNR1 Putative uncharacterized protein At5g61030 n=1 ... 151 4e-35
UniRef100_Q2QQ97 Os12g0502200 protein n=1 Tax=Oryza sativa Japon... 150 5e-35
UniRef100_B1Q4T1 Glycine-rich protein n=1 Tax=Bruguiera gymnorhi... 150 5e-35
UniRef100_Q8S2Y0 Glycine-rich RNA binding protein (Fragment) n=1... 150 7e-35
UniRef100_Q8S2X5 Glycine-rich RNA binding protein (Fragment) n=1... 150 7e-35
UniRef100_Q8RUC2 Glycine-rich RNA binding protein (Fragment) n=1... 150 7e-35
UniRef100_B8A3G8 Putative uncharacterized protein n=1 Tax=Zea ma... 150 7e-35
UniRef100_B0D8X8 Predicted protein n=1 Tax=Laccaria bicolor S238... 150 9e-35
UniRef100_C1ZIK1 RRM domain-containing RNA-binding protein n=1 T... 149 2e-34
UniRef100_C6SV67 Putative uncharacterized protein n=1 Tax=Glycin... 149 2e-34
UniRef100_A2BZB6 RNA-binding region RNP-1 (RNA recognition motif... 148 3e-34
UniRef100_B9HRM9 Predicted protein (Fragment) n=1 Tax=Populus tr... 148 3e-34
UniRef100_A7PDA4 Chromosome chr17 scaffold_12, whole genome shot... 148 3e-34
UniRef100_A5B074 Putative uncharacterized protein n=1 Tax=Vitis ... 148 3e-34
UniRef100_C0H8U7 Cold-inducible RNA-binding protein n=1 Tax=Salm... 147 4e-34
UniRef100_Q8S2Y8 Glycine-rich RNA binding protein (Fragment) n=7... 147 4e-34
UniRef100_Q8S2Y7 Glycine-rich RNA binding protein (Fragment) n=1... 147 4e-34
UniRef100_Q8S2X6 Glycine-rich RNA binding protein (Fragment) n=1... 147 4e-34
UniRef100_A2BTV1 RNA-binding region RNP-1 (RNA recognition motif... 147 7e-34
UniRef100_A9B9L1 RNA-binding region RNP-1 (RNA recognition motif... 146 1e-33
UniRef100_A7EC48 Glycine-rich RNA-binding protein n=1 Tax=Sclero... 146 1e-33
UniRef100_Q7VEJ9 RNA-binding protein, RRM domain n=1 Tax=Prochlo... 145 2e-33
UniRef100_B6T5N8 Glycine-rich RNA-binding protein 2 n=1 Tax=Zea ... 145 2e-33
UniRef100_Q8S2Y4 Glycine-rich RNA binding protein (Fragment) n=1... 145 2e-33
UniRef100_Q8S2Y5 Glycine-rich RNA binding protein (Fragment) n=1... 145 3e-33
UniRef100_C5YNX7 Putative uncharacterized protein Sb08g015580 n=... 145 3e-33
UniRef100_A5GHM5 RNA-binding protein, RRM domain n=1 Tax=Synecho... 144 5e-33
UniRef100_A2C5L0 RNA-binding region RNP-1 (RNA recognition motif... 144 5e-33
UniRef100_A4CWS8 RNA-binding region RNP-1 (RNA recognition motif... 144 5e-33
UniRef100_Q7UA83 RNA-binding region RNP-1 (RNA recognition motif... 143 1e-32
UniRef100_A2SMG9 RNA-binding region RNP-1 n=1 Tax=Methylibium pe... 142 1e-32
UniRef100_Q062H8 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 142 1e-32
UniRef100_C5XDU4 Putative uncharacterized protein Sb02g038590 n=... 142 1e-32
UniRef100_A5GPV8 RNA-binding protein, RRM domain n=1 Tax=Synecho... 142 2e-32
UniRef100_B4F8A7 Putative uncharacterized protein n=1 Tax=Zea ma... 142 2e-32
UniRef100_Q46I49 RNA-binding region RNP-1 (RNA recognition motif... 142 2e-32
UniRef100_Q9M6A1 Putative glycine-rich RNA binding protein 1 n=1... 141 3e-32
UniRef100_C4J159 Putative uncharacterized protein n=1 Tax=Zea ma... 141 3e-32
UniRef100_A8IZS5 Glycine-rich RNA-binding protein n=1 Tax=Chlamy... 141 3e-32
UniRef100_A7SMB3 Predicted protein n=1 Tax=Nematostella vectensi... 141 3e-32
UniRef100_Q8S2X7 Glycine-rich RNA binding protein (Fragment) n=2... 141 4e-32
UniRef100_Q9U5Y7 Glycine rich RNA binding protein n=1 Tax=Ciona ... 140 5e-32
UniRef100_Q31DH1 RNA-binding region RNP-1 n=1 Tax=Prochlorococcu... 140 7e-32
UniRef100_Q9XY11 Cold-inducible RNA-binding protein n=1 Tax=Cion... 140 7e-32
UniRef100_Q7UEZ2 RNA-binding protein n=1 Tax=Rhodopirellula balt... 140 9e-32
UniRef100_Q8VXC4 Glycine rich RNA binding protein n=1 Tax=Oryza ... 140 9e-32
UniRef100_A6RXS7 Putative uncharacterized protein n=1 Tax=Botryo... 140 9e-32
UniRef100_Q7V9D6 RNA-binding region RNP-1 (RNA recognition motif... 139 2e-31
UniRef100_A7SMA3 Predicted protein n=1 Tax=Nematostella vectensi... 139 2e-31
UniRef100_A8G200 RNA-binding region RNP-1 (RNA recognition motif... 139 2e-31
UniRef100_A6GBL5 RNA-binding protein n=1 Tax=Plesiocystis pacifi... 139 2e-31
UniRef100_B2WDB7 Putative uncharacterized protein n=1 Tax=Pyreno... 139 2e-31
UniRef100_A2ZKT5 Putative uncharacterized protein n=1 Tax=Oryza ... 138 3e-31
UniRef100_B6UB95 Glycine-rich RNA-binding protein 7 n=1 Tax=Zea ... 137 4e-31
UniRef100_Q9SIX3 Putative glycine-rich RNA-binding protein n=1 T... 137 6e-31
UniRef100_C3Y7T8 Putative uncharacterized protein n=1 Tax=Branch... 137 6e-31
UniRef100_Q566W6 Zgc:112425 n=1 Tax=Danio rerio RepID=Q566W6_DANRE 136 1e-30
UniRef100_A3PA60 RNA-binding region RNP-1 (RNA recognition motif... 136 1e-30
UniRef100_A8HQ72 SR protein factor n=1 Tax=Chlamydomonas reinhar... 136 1e-30
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 136 1e-30
UniRef100_B5DGC5 Cold-inducible RNA-binding protein n=1 Tax=Salm... 136 1e-30
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 136 1e-30
UniRef100_B6TI13 Glycine-rich RNA-binding protein 7 n=1 Tax=Zea ... 135 3e-30
UniRef100_B5X2C8 Cold-inducible RNA-binding protein n=1 Tax=Salm... 134 4e-30
UniRef100_B5DGP8 Elongation factor-1 delta-1 n=1 Tax=Salmo salar... 134 4e-30
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 134 5e-30
UniRef100_A1WE85 RNP-1 like RNA-binding protein n=1 Tax=Verminep... 134 6e-30
UniRef100_B4FFS8 Putative uncharacterized protein n=1 Tax=Zea ma... 133 8e-30
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 133 1e-29
UniRef100_B5RI95 Cold-inducible RNA-binding protein n=1 Tax=Salm... 133 1e-29
UniRef100_A3ZRX9 RNA-binding protein n=1 Tax=Blastopirellula mar... 133 1e-29
UniRef100_B6T255 Putative uncharacterized protein n=1 Tax=Zea ma... 133 1e-29
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 133 1e-29
UniRef100_A7SMA7 Predicted protein n=1 Tax=Nematostella vectensi... 133 1e-29
UniRef100_B9EN93 Cold-inducible RNA-binding protein n=1 Tax=Salm... 132 1e-29
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 132 1e-29
UniRef100_Q0UUY3 Putative uncharacterized protein n=1 Tax=Phaeos... 132 1e-29
UniRef100_A8N6M0 Putative uncharacterized protein n=1 Tax=Coprin... 132 1e-29
UniRef100_UPI0001745B20 RNA-binding region RNP-1 n=1 Tax=Verruco... 132 2e-29
UniRef100_B4F7T1 Putative uncharacterized protein n=1 Tax=Zea ma... 132 2e-29
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 132 2e-29
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 132 2e-29
UniRef100_UPI00016C5334 RNA-binding region RNP-1 n=1 Tax=Gemmata... 131 3e-29
UniRef100_C6TJN7 Putative uncharacterized protein n=1 Tax=Glycin... 131 3e-29
UniRef100_C0PEX7 Putative uncharacterized protein n=1 Tax=Zea ma... 131 3e-29
UniRef100_A1TWH4 RNP-1-like RNA-binding protein n=1 Tax=Acidovor... 131 4e-29
UniRef100_B6TR84 Glycine-rich RNA-binding protein 7 n=1 Tax=Zea ... 131 4e-29
UniRef100_A9EYW0 RNA-binding region RNP-1 (RNA recognition motif... 130 5e-29
UniRef100_C1BH84 Cold-inducible RNA-binding protein n=1 Tax=Onco... 130 7e-29
UniRef100_A7T8T6 Predicted protein n=1 Tax=Nematostella vectensi... 130 7e-29
UniRef100_UPI00016E9D6D UPI00016E9D6D related cluster n=1 Tax=Ta... 130 9e-29
UniRef100_Q05UU1 RNA-binding region RNP-1 (RNA recognition motif... 130 9e-29
UniRef100_A9RSY7 Predicted protein n=1 Tax=Physcomitrella patens... 129 1e-28
UniRef100_Q3B0Y9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 129 2e-28
UniRef100_Q5KDS6 Glycine-rich RNA binding protein, putative n=1 ... 129 2e-28
UniRef100_UPI0000588CC7 PREDICTED: similar to MGC68480 protein n... 129 2e-28
UniRef100_Q7V3Q7 RNA-binding region RNP-1 (RNA recognition motif... 129 2e-28
UniRef100_B1Y6Y3 RNP-1 like RNA-binding protein n=1 Tax=Leptothr... 129 2e-28
UniRef100_B5X3A3 Cold-inducible RNA-binding protein n=1 Tax=Salm... 128 3e-28
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 128 3e-28
UniRef100_Q7ZUQ9 Cold inducible RNA binding protein n=1 Tax=Dani... 128 4e-28
UniRef100_Q1LVU9 Cold inducible RNA binding protein (Fragment) n... 128 4e-28
UniRef100_Q1LVU8 Cold inducible RNA binding protein (Fragment) n... 128 4e-28
UniRef100_C1BJ24 Cold-inducible RNA-binding protein n=1 Tax=Osme... 128 4e-28
UniRef100_Q109W0 Os10g0321700 protein n=1 Tax=Oryza sativa Japon... 128 4e-28
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 127 5e-28
UniRef100_UPI00016C3DAE RNA-binding region RNP-1 n=1 Tax=Gemmata... 127 5e-28
UniRef100_Q9C909 Putative RNA-binding protein; 37609-36098 n=1 T... 127 5e-28
UniRef100_Q7SGB3 Predicted protein n=1 Tax=Neurospora crassa Rep... 127 6e-28
UniRef100_UPI0001927785 PREDICTED: similar to cold inducible RNA... 126 1e-27
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 126 1e-27
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 126 1e-27
UniRef100_UPI00017466C4 RNA-binding protein (RRM domain) n=1 Tax... 125 2e-27
UniRef100_A9C1L2 RNP-1 like RNA-binding protein n=1 Tax=Delftia ... 125 2e-27
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 125 2e-27
UniRef100_A9NNN7 Putative uncharacterized protein n=1 Tax=Picea ... 125 2e-27
UniRef100_Q5KDS5 Glycine-rich RNA binding protein, putative n=1 ... 125 2e-27
UniRef100_C0H8N3 Heterogeneous nuclear ribonucleoprotein A1 n=1 ... 125 3e-27
UniRef100_B9MIH3 RNP-1 like RNA-binding protein n=1 Tax=Diaphoro... 125 3e-27
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 125 3e-27
UniRef100_Q75LJ7 Os03g0836200 protein n=2 Tax=Oryza sativa RepID... 125 3e-27
UniRef100_Q0IE69 RNA-binding region RNP-1 (RNA recognition motif... 124 4e-27
UniRef100_C6TEW4 Putative uncharacterized protein n=1 Tax=Glycin... 124 5e-27
UniRef100_C1MIF9 Predicted protein n=1 Tax=Micromonas pusilla CC... 124 5e-27
UniRef100_C1E9S3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 124 5e-27
UniRef100_Q7ZYV6 Hyperosmotic glycine rich protein n=1 Tax=Salmo... 124 7e-27
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 124 7e-27
UniRef100_A9NQT3 Putative uncharacterized protein n=1 Tax=Picea ... 124 7e-27
UniRef100_B7QN34 Putative uncharacterized protein (Fragment) n=1... 124 7e-27
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 123 9e-27
UniRef100_Q3MC83 Putative uncharacterized protein n=1 Tax=Anabae... 123 9e-27
UniRef100_Q9SP10 Glycine-rich RNA binding protein (Fragment) n=1... 123 9e-27
UniRef100_B4R5S1 GD17272 n=1 Tax=Drosophila simulans RepID=B4R5S... 123 1e-26
UniRef100_Q941H8 RNA-binding protein n=1 Tax=Solanum tuberosum R... 122 1e-26
UniRef100_B7PDJ6 Putative uncharacterized protein (Fragment) n=1... 122 1e-26
UniRef100_UPI000186773D hypothetical protein BRAFLDRAFT_237208 n... 122 2e-26
UniRef100_UPI00016C4475 putative RNA-binding protein n=1 Tax=Gem... 122 2e-26
UniRef100_UPI0001867639 hypothetical protein BRAFLDRAFT_237268 n... 122 3e-26
UniRef100_UPI0000DA3D7B PREDICTED: hypothetical protein n=1 Tax=... 122 3e-26
UniRef100_D0CMI9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 122 3e-26
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 122 3e-26
UniRef100_Q3E9N5 Putative uncharacterized protein At4g39260.4 n=... 122 3e-26
UniRef100_C0PNT1 Putative uncharacterized protein n=1 Tax=Zea ma... 122 3e-26
UniRef100_A0NCZ0 AGAP001055-PA (Fragment) n=1 Tax=Anopheles gamb... 122 3e-26
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 121 3e-26
UniRef100_A2BND9 RNA-binding region RNP-1 (RNA recognition motif... 121 3e-26
UniRef100_B7PFE5 Secreted protein, putative (Fragment) n=1 Tax=I... 121 3e-26
UniRef100_B4MID6 GK23346 n=1 Tax=Drosophila willistoni RepID=B4M... 121 3e-26
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 121 4e-26
UniRef100_Q8JJB1 Cold inducible RNA-binding protein beta n=1 Tax... 121 4e-26
UniRef100_Q3ANN6 RNA-binding region RNP-1 (RNA recognition motif... 121 4e-26
UniRef100_A1VWW4 RNP-1 like RNA-binding protein n=1 Tax=Polaromo... 121 4e-26
UniRef100_Q3E9N6 Putative uncharacterized protein At4g39260.3 n=... 121 4e-26
UniRef100_B7PCR0 Cement protein, putative (Fragment) n=1 Tax=Ixo... 121 4e-26
UniRef100_Q9SVM8 Glycine-rich RNA-binding protein 2, mitochondri... 121 4e-26
UniRef100_UPI00015B5EB5 PREDICTED: similar to CG3606-PB n=1 Tax=... 120 6e-26
UniRef100_Q5TU53 AGAP002965-PA (Fragment) n=1 Tax=Anopheles gamb... 120 6e-26
UniRef100_B7PE98 Glycine rich protein, putative (Fragment) n=1 T... 120 7e-26
UniRef100_B7P671 Cement protein, putative n=1 Tax=Ixodes scapula... 120 7e-26
UniRef100_A7SMB0 Predicted protein n=1 Tax=Nematostella vectensi... 120 7e-26
UniRef100_UPI0001BAFCB9 RNP-1 like RNA-binding protein n=1 Tax=H... 120 1e-25
UniRef100_UPI0001925556 PREDICTED: similar to Putative RNA-bindi... 120 1e-25
UniRef100_UPI00016E78CD UPI00016E78CD related cluster n=1 Tax=Ta... 120 1e-25
UniRef100_Q121M2 RNA-binding region RNP-1 (RNA recognition motif... 120 1e-25
UniRef100_C5CY78 RNP-1 like RNA-binding protein n=1 Tax=Variovor... 120 1e-25
UniRef100_Q804J2 Nucleolin n=1 Tax=Cyprinus carpio RepID=Q804J2_... 119 1e-25
UniRef100_B7QAD8 Secreted protein, putative (Fragment) n=1 Tax=I... 119 1e-25
UniRef100_B7PFE7 Cement protein, putative (Fragment) n=1 Tax=Ixo... 119 1e-25
UniRef100_B3NX66 GG17948 n=1 Tax=Drosophila erecta RepID=B3NX66_... 119 1e-25
UniRef100_UPI000186D45B RNA-binding protein cabeza, putative n=1... 119 2e-25
UniRef100_Q7SXQ3 Zgc:66127 n=1 Tax=Danio rerio RepID=Q7SXQ3_DANRE 119 2e-25
UniRef100_C4JAE0 Putative uncharacterized protein n=1 Tax=Zea ma... 119 2e-25
UniRef100_A9SWE4 Predicted protein n=1 Tax=Physcomitrella patens... 119 2e-25
UniRef100_UPI0000E491FD PREDICTED: hypothetical protein, partial... 119 2e-25
UniRef100_UPI0000DB7202 PREDICTED: hypothetical protein n=1 Tax=... 119 2e-25
UniRef100_UPI000069F523 Heterogeneous nuclear ribonucleoprotein ... 118 3e-25
UniRef100_B0BM91 LOC100144993 protein n=1 Tax=Xenopus (Silurana)... 118 3e-25
UniRef100_Q941H9 RNA-binding protein n=1 Tax=Nicotiana tabacum R... 118 3e-25
UniRef100_Q7XI13 Os07g0187300 protein n=1 Tax=Oryza sativa Japon... 118 3e-25
UniRef100_B8B7X7 Putative uncharacterized protein n=1 Tax=Oryza ... 118 3e-25
UniRef100_B7P0W1 Cement protein, putative (Fragment) n=1 Tax=Ixo... 118 3e-25
UniRef100_UPI0000DA1BD4 PREDICTED: hypothetical protein n=1 Tax=... 118 4e-25
UniRef100_UPI0000025476 DEAH (Asp-Glu-Ala-His) box polypeptide 9... 118 4e-25
UniRef100_UPI00001EDBAC DEAH (Asp-Glu-Ala-His) box polypeptide 9... 118 4e-25
UniRef100_B6ID90 DEAH (Asp-Glu-Ala-His) box polypeptide 9 (Fragm... 118 4e-25
UniRef100_UPI0001A2D182 heterogeneous nuclear ribonucleoprotein ... 117 5e-25
UniRef100_Q6PHJ4 Hnrpa0 protein (Fragment) n=1 Tax=Danio rerio R... 117 5e-25
UniRef100_Q6NYW8 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 117 5e-25
UniRef100_A1VW19 RNP-1 like RNA-binding protein n=1 Tax=Polaromo... 117 5e-25
UniRef100_C5XYF2 Putative uncharacterized protein Sb04g007860 n=... 117 5e-25
UniRef100_A9SV56 Predicted protein (Fragment) n=1 Tax=Physcomitr... 117 5e-25
UniRef100_B7P9A9 HyFMR1 protein, putative (Fragment) n=1 Tax=Ixo... 117 5e-25
UniRef100_B4JJY5 GH12560 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 117 5e-25
UniRef100_Q2V614 Putative uncharacterized protein n=1 Tax=Brassi... 106 5e-25
UniRef100_Q7T050 Nucleolin 2 n=1 Tax=Cyprinus carpio RepID=Q7T05... 117 6e-25
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 117 6e-25
UniRef100_C5MJJ9 Predicted protein n=1 Tax=Candida tropicalis MY... 117 6e-25
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 117 8e-25
UniRef100_B7ZR96 LOC397919 protein n=1 Tax=Xenopus laevis RepID=... 117 8e-25
UniRef100_A1VUR7 RNP-1 like RNA-binding protein n=1 Tax=Polaromo... 117 8e-25
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 117 8e-25
UniRef100_C0HED8 Putative uncharacterized protein n=1 Tax=Zea ma... 117 8e-25
UniRef100_UPI00015539C6 PREDICTED: hypothetical protein n=1 Tax=... 116 1e-24
UniRef100_UPI00016E9D6B UPI00016E9D6B related cluster n=1 Tax=Ta... 116 1e-24
UniRef100_Q06459 Nucleolin n=1 Tax=Xenopus laevis RepID=Q06459_X... 116 1e-24
UniRef100_B7ZR98 LOC397919 protein n=1 Tax=Xenopus laevis RepID=... 116 1e-24
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 116 1e-24
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 116 1e-24
UniRef100_C6TK08 Putative uncharacterized protein n=1 Tax=Glycin... 116 1e-24
UniRef100_A9PIG6 Putative uncharacterized protein n=1 Tax=Populu... 116 1e-24
UniRef100_A2X2I9 Putative uncharacterized protein n=1 Tax=Oryza ... 116 1e-24
UniRef100_P51992 Heterogeneous nuclear ribonucleoprotein A3 homo... 116 1e-24
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 116 1e-24
UniRef100_UPI000069F524 Heterogeneous nuclear ribonucleoprotein ... 116 1e-24
UniRef100_A1WDA4 RNP-1 like RNA-binding protein n=1 Tax=Acidovor... 116 1e-24
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 116 1e-24
UniRef100_Q8YQB7 All3916 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 115 2e-24
UniRef100_B9R282 'Cold-shock' DNA-binding domain protein n=1 Tax... 115 2e-24
UniRef100_Q6YVH4 Putative heterogeneous nuclearribonucleoprotein... 115 2e-24
UniRef100_Q40436 RNA-binding glycine rich protein n=1 Tax=Nicoti... 115 2e-24
UniRef100_B9S651 RNA binding protein, putative n=1 Tax=Ricinus c... 115 2e-24
UniRef100_B4IF47 GM13418 n=1 Tax=Drosophila sechellia RepID=B4IF... 115 2e-24
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 115 2e-24
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 115 2e-24
UniRef100_Q6YVH3 Os02g0221500 protein n=2 Tax=Oryza sativa Japon... 115 2e-24
UniRef100_B9PV72 Putative uncharacterized protein n=1 Tax=Toxopl... 115 2e-24
UniRef100_B7PYQ7 Putative uncharacterized protein (Fragment) n=1... 115 2e-24
UniRef100_B6GW69 Pc06g01290 protein n=1 Tax=Penicillium chrysoge... 115 2e-24
UniRef100_Q7T049 Nucleolin n=1 Tax=Cyprinus carpio RepID=Q7T049_... 115 3e-24
UniRef100_C1V3H1 RRM domain-containing RNA-binding protein n=1 T... 115 3e-24
UniRef100_B7ZQP4 Putative uncharacterized protein n=1 Tax=Xenopu... 114 4e-24
UniRef100_B9NZ72 RNA-binding region RNP-1 (RNA recognition motif... 114 4e-24
UniRef100_Q42412 RNA-binding protein RZ-1 n=1 Tax=Nicotiana sylv... 114 4e-24
UniRef100_B9SNV6 Glycine-rich cell wall structural protein 1.8, ... 114 4e-24
UniRef100_B6TGB9 Heterogeneous nuclear ribonucleoprotein A3 n=1 ... 114 4e-24
UniRef100_UPI00015539BB PREDICTED: hypothetical protein n=1 Tax=... 114 5e-24
UniRef100_Q11SF1 RNA binding protein n=1 Tax=Cytophaga hutchinso... 114 5e-24
UniRef100_Q0D4V3 Os07g0602600 protein n=1 Tax=Oryza sativa Japon... 114 5e-24
UniRef100_C4J0A5 Putative uncharacterized protein n=1 Tax=Zea ma... 114 5e-24
UniRef100_B6TF12 Glycine-rich RNA-binding protein 8 n=1 Tax=Zea ... 114 5e-24
UniRef100_B4FHJ8 Putative uncharacterized protein n=1 Tax=Zea ma... 114 5e-24
UniRef100_B9QIA3 Putative uncharacterized protein n=1 Tax=Toxopl... 114 5e-24
UniRef100_B6KPI0 Putative uncharacterized protein n=1 Tax=Toxopl... 114 5e-24
UniRef100_P51968 Heterogeneous nuclear ribonucleoprotein A3 homo... 114 5e-24
UniRef100_Q27294 RNA-binding protein cabeza n=1 Tax=Drosophila m... 114 5e-24
UniRef100_B5XF66 Cold-inducible RNA-binding protein n=1 Tax=Salm... 114 7e-24
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 114 7e-24
UniRef100_B1JXE0 Putative uncharacterized protein n=1 Tax=Burkho... 114 7e-24
UniRef100_O93465 RNA-binding protein AxRNBP n=1 Tax=Ambystoma me... 113 9e-24
UniRef100_B4MNL3 GK19601 n=1 Tax=Drosophila willistoni RepID=B4M... 113 9e-24
UniRef100_B9RLN6 Glycine-rich RNA-binding protein, putative n=1 ... 113 1e-23
UniRef100_B7Q768 Secreted protein, putative (Fragment) n=1 Tax=I... 113 1e-23
UniRef100_C5MJK0 Predicted protein n=1 Tax=Candida tropicalis MY... 113 1e-23
UniRef100_O70133-2 Isoform 2 of ATP-dependent RNA helicase A n=1... 113 1e-23
UniRef100_O70133 ATP-dependent RNA helicase A n=2 Tax=Mus muscul... 113 1e-23
UniRef100_UPI0001B7A53E UPI0001B7A53E related cluster n=1 Tax=Ra... 112 2e-23
UniRef100_UPI00016E9D6C UPI00016E9D6C related cluster n=1 Tax=Ta... 112 2e-23
UniRef100_Q8JJB2 Cold inducible RNA-binding protein alpha n=1 Ta... 112 2e-23
UniRef100_B7Q3T8 Putative uncharacterized protein (Fragment) n=1... 112 2e-23
UniRef100_UPI00016C4C95 RNA-binding protein n=1 Tax=Gemmata obsc... 112 2e-23
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 112 2e-23
UniRef100_B9EP65 Cold-inducible RNA-binding protein n=1 Tax=Salm... 112 2e-23
UniRef100_A9G206 RNA-binding protein n=1 Tax=Sorangium cellulosu... 112 2e-23
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 112 2e-23
UniRef100_C6T2P6 Putative uncharacterized protein n=1 Tax=Glycin... 112 2e-23
UniRef100_A4S1A8 Predicted protein n=1 Tax=Ostreococcus lucimari... 112 2e-23
UniRef100_B4PQG9 GE24426 n=1 Tax=Drosophila yakuba RepID=B4PQG9_... 112 2e-23
UniRef100_UPI00016C4BFC hypothetical protein GobsU_21505 n=1 Tax... 112 3e-23
UniRef100_UPI0000E4714A PREDICTED: hypothetical protein, partial... 112 3e-23
UniRef100_B2RXM2 EG627828 protein n=1 Tax=Mus musculus RepID=B2R... 112 3e-23
UniRef100_Q39337 Glycine-rich_protein_(Aa1-291) n=1 Tax=Brassica... 112 3e-23
UniRef100_B8B8D6 Putative uncharacterized protein n=1 Tax=Oryza ... 112 3e-23
UniRef100_A3BDN5 Putative uncharacterized protein n=1 Tax=Oryza ... 112 3e-23
UniRef100_B1X5L4 RNA-binding protein RbpD n=1 Tax=Paulinella chr... 112 3e-23
UniRef100_B0S6Z1 Novel protein similar to vertebrate DEAH (Asp-G... 111 3e-23
UniRef100_A9GC81 RNA-binding protein n=1 Tax=Sorangium cellulosu... 111 3e-23
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 111 3e-23
UniRef100_Q3EA40 Putative uncharacterized protein At4g13850.2 n=... 111 3e-23
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 111 3e-23
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 111 3e-23
UniRef100_B7Q8C0 Secreted protein, putative (Fragment) n=1 Tax=I... 111 3e-23
UniRef100_B4MGC4 GJ18490 n=1 Tax=Drosophila virilis RepID=B4MGC4... 111 3e-23
UniRef100_B3N0U3 GF19034 n=1 Tax=Drosophila ananassae RepID=B3N0... 111 3e-23
UniRef100_C9J285 Putative uncharacterized protein ENSP0000039240... 111 3e-23
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 111 3e-23
UniRef100_UPI000155FB50 PREDICTED: similar to heterogeneous nucl... 111 4e-23
UniRef100_UPI00004289ED PREDICTED: similar to heterogeneous nucl... 111 4e-23
UniRef100_UPI0000194843 PREDICTED: similar to heterogeneous nucl... 111 4e-23
UniRef100_UPI00015DF065 UPI00015DF065 related cluster n=1 Tax=Mu... 111 4e-23
UniRef100_Q9FM47 Similarity to RNA binding protein n=1 Tax=Arabi... 111 4e-23
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 111 4e-23
UniRef100_B5DYF4 GA26796 n=1 Tax=Drosophila pseudoobscura pseudo... 111 4e-23
UniRef100_UPI000186F0E7 RNA-binding protein squid, putative n=1 ... 110 6e-23
UniRef100_UPI00017C2E1B PREDICTED: similar to heterogeneous nucl... 110 6e-23
UniRef100_UPI0000F2EC55 PREDICTED: similar to heterogeneous nucl... 110 6e-23
UniRef100_UPI00005A58F4 PREDICTED: similar to heterogeneous nucl... 110 6e-23
UniRef100_UPI0001B7A586 regulator of G-protein signaling like 1 ... 110 6e-23
UniRef100_UPI0000EBC230 PREDICTED: Bos taurus hypothetical prote... 110 6e-23
UniRef100_Q43522 Tfm5 protein n=1 Tax=Solanum lycopersicum RepID... 110 6e-23
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 110 6e-23
UniRef100_B9HFC9 Predicted protein n=1 Tax=Populus trichocarpa R... 110 6e-23
UniRef100_A9NJZ7 Putative uncharacterized protein n=1 Tax=Picea ... 110 6e-23
UniRef100_B4NEW8 GK25719 n=1 Tax=Drosophila willistoni RepID=B4N... 110 6e-23
UniRef100_UPI0001982DB4 PREDICTED: hypothetical protein n=1 Tax=... 110 8e-23
UniRef100_UPI0000E4626F PREDICTED: similar to heterogeneous nucl... 110 8e-23
UniRef100_UPI0000DA2377 PREDICTED: hypothetical protein n=1 Tax=... 110 8e-23
UniRef100_UPI0000DA1CBA PREDICTED: hypothetical protein n=1 Tax=... 110 8e-23
UniRef100_UPI00005A58F9 PREDICTED: similar to heterogeneous nucl... 110 8e-23
UniRef100_UPI00005A58F2 PREDICTED: similar to heterogeneous nucl... 110 8e-23
UniRef100_UPI00015A499E nucleolin n=1 Tax=Danio rerio RepID=UPI0... 110 8e-23
UniRef100_UPI0000568F42 heterogeneous nuclear ribonucleoprotein ... 110 8e-23
UniRef100_Q803K3 Heterogeneous nuclear ribonucleoprotein A1 n=1 ... 110 8e-23
UniRef100_Q7ZX33 Hnrpa1 protein n=1 Tax=Xenopus laevis RepID=Q7Z... 110 8e-23
UniRef100_Q08CQ9 Zgc:152810 n=1 Tax=Danio rerio RepID=Q08CQ9_DANRE 110 8e-23
UniRef100_C1BM74 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 110 8e-23
UniRef100_B8JLQ3 Novel protein similar to H.sapiens NCL, nucleol... 110 8e-23
UniRef100_B3DH54 Zgc:152810 protein n=1 Tax=Danio rerio RepID=B3... 110 8e-23
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 110 8e-23
UniRef100_A0NYD4 Cold shock protein n=1 Tax=Labrenzia aggregata ... 110 8e-23
UniRef100_Q00RR6 H0525G02.4 protein n=2 Tax=Oryza sativa RepID=Q... 110 8e-23
UniRef100_B7QMH8 H/ACA ribonucleoprotein complex protein, putati... 110 8e-23
UniRef100_P21522 Heterogeneous nuclear ribonucleoprotein A1, A2/... 110 8e-23
UniRef100_Q7XTS4 Protein argonaute 2 n=1 Tax=Oryza sativa Japoni... 110 8e-23
UniRef100_UPI0001757FBF PREDICTED: similar to T19B10.5 n=1 Tax=T... 110 1e-22
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 110 1e-22
UniRef100_UPI0000D67C8E PREDICTED: similar to heterogeneous nucl... 110 1e-22
UniRef100_UPI0000429BC9 PREDICTED: similar to heterogeneous nucl... 110 1e-22
UniRef100_UPI0000222BCC Hypothetical protein CBG04553 n=1 Tax=Ca... 110 1e-22
UniRef100_Q3UZG3 Putative uncharacterized protein n=1 Tax=Mus mu... 110 1e-22
UniRef100_Q0VG47 Heterogeneous nuclear ribonucleoprotein A3 n=5 ... 110 1e-22
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 110 1e-22
UniRef100_C0P8S9 Putative uncharacterized protein n=1 Tax=Zea ma... 110 1e-22
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 110 1e-22
UniRef100_B5DW02 GA27272 n=1 Tax=Drosophila pseudoobscura pseudo... 110 1e-22
UniRef100_B4L7Q3 GI11213 n=1 Tax=Drosophila mojavensis RepID=B4L... 110 1e-22
UniRef100_Q8BG05 Heterogeneous nuclear ribonucleoprotein A3 n=3 ... 110 1e-22
UniRef100_UPI000155D044 PREDICTED: similar to heterogeneous nucl... 109 1e-22
UniRef100_UPI0001553727 PREDICTED: similar to heterogeneous nucl... 109 1e-22
UniRef100_UPI0000E80910 PREDICTED: heterogeneous nuclear ribonuc... 109 1e-22
UniRef100_UPI00005A58FB PREDICTED: similar to heterogeneous nucl... 109 1e-22
UniRef100_Q5Z4P5 Os06g0317400 protein n=1 Tax=Oryza sativa Japon... 109 1e-22
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 109 1e-22
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 109 1e-22
UniRef100_B4N8P2 GK12096 n=1 Tax=Drosophila willistoni RepID=B4N... 109 1e-22
UniRef100_B4HFQ5 GM25918 n=1 Tax=Drosophila sechellia RepID=B4HF... 109 1e-22
UniRef100_B4G3A6 GL23478 n=1 Tax=Drosophila persimilis RepID=B4G... 109 1e-22
UniRef100_B2B6T1 Predicted CDS Pa_2_8940 n=1 Tax=Podospora anser... 109 1e-22
UniRef100_UPI0000DB6D77 PREDICTED: similar to CG5913-PA n=1 Tax=... 109 2e-22
UniRef100_UPI00005A58FC PREDICTED: similar to heterogeneous nucl... 109 2e-22
UniRef100_Q1BU84 Putative uncharacterized protein n=1 Tax=Burkho... 109 2e-22
UniRef100_Q69XV3 Os06g0622400 protein n=1 Tax=Oryza sativa Japon... 109 2e-22
UniRef100_Q5Z655 Os06g0566100 protein n=3 Tax=Oryza sativa Japon... 109 2e-22
UniRef100_A2YE63 Putative uncharacterized protein n=1 Tax=Oryza ... 109 2e-22
UniRef100_C8VV67 RE34829p n=2 Tax=Drosophila melanogaster RepID=... 109 2e-22
UniRef100_B4QYR3 GD21374 n=1 Tax=Drosophila simulans RepID=B4QYR... 109 2e-22
UniRef100_B4IH20 GM16385 n=1 Tax=Drosophila sechellia RepID=B4IH... 109 2e-22
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 109 2e-22
UniRef100_A4V3J6 Heterogeneous nuclear ribonucleoprotein at 98DE... 109 2e-22
UniRef100_P07909-2 Isoform A of Heterogeneous nuclear ribonucleo... 109 2e-22
UniRef100_P07909 Heterogeneous nuclear ribonucleoprotein A1 n=2 ... 109 2e-22
UniRef100_UPI0000DB6C07 PREDICTED: hypothetical protein n=1 Tax=... 108 2e-22
UniRef100_UPI00005A58F8 PREDICTED: similar to heterogeneous nucl... 108 2e-22
UniRef100_UPI0001B7B4B4 UPI0001B7B4B4 related cluster n=1 Tax=Ra... 108 2e-22
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 108 2e-22
UniRef100_O65514 Putative glycine-rich cell wall protein n=1 Tax... 108 2e-22
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 108 2e-22
UniRef100_C5XP48 Putative uncharacterized protein Sb03g005056 (F... 108 2e-22
UniRef100_A9NNT8 Putative uncharacterized protein n=1 Tax=Picea ... 108 2e-22
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 108 2e-22
UniRef100_A5BQ96 Putative uncharacterized protein n=1 Tax=Vitis ... 108 2e-22
UniRef100_UPI0000E127A2 Os06g0317400 n=1 Tax=Oryza sativa Japoni... 108 3e-22
UniRef100_UPI0000DA36CE PREDICTED: hypothetical protein n=1 Tax=... 108 3e-22
UniRef100_Q6NYB0 Zgc:77366 n=1 Tax=Danio rerio RepID=Q6NYB0_DANRE 108 3e-22
UniRef100_B5X0T7 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 108 3e-22
UniRef100_Q0BCI9 Putative uncharacterized protein n=1 Tax=Burkho... 108 3e-22
UniRef100_Q9SIH2 Putative uncharacterized protein At2g36120 n=1 ... 108 3e-22
UniRef100_Q7Y218 Putative uncharacterized protein At5g46730 n=1 ... 108 3e-22
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 108 3e-22
UniRef100_Q8I519 Erythrocyte membrane protein 1, PfEMP1 n=1 Tax=... 108 3e-22
UniRef100_B6KME4 RRM domain-containing protein n=1 Tax=Toxoplasm... 108 3e-22
UniRef100_B4R7C9 GD16486 n=1 Tax=Drosophila simulans RepID=B4R7C... 108 3e-22
UniRef100_B4NBV5 GK25731 n=1 Tax=Drosophila willistoni RepID=B4N... 108 3e-22
UniRef100_A8XS85 C. briggsae CBR-DRR-2 protein n=1 Tax=Caenorhab... 108 3e-22
UniRef100_P51991-2 Isoform 2 of Heterogeneous nuclear ribonucleo... 108 3e-22
UniRef100_P51991 Heterogeneous nuclear ribonucleoprotein A3 n=1 ... 108 3e-22
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 108 4e-22
UniRef100_UPI00017931E0 PREDICTED: similar to heterogeneous nucl... 108 4e-22
UniRef100_UPI0001552E25 PREDICTED: hypothetical protein n=1 Tax=... 108 4e-22
UniRef100_UPI00004367E2 hypothetical protein LOC323529 n=1 Tax=D... 108 4e-22
UniRef100_UPI0000ECB6DA UPI0000ECB6DA related cluster n=1 Tax=Ga... 108 4e-22
UniRef100_UPI0000ECAE31 Heterogeneous nuclear ribonucleoprotein ... 108 4e-22
UniRef100_Q9DFG5 Cold-inducible RNA binding protein XCIRP-1 n=1 ... 108 4e-22
UniRef100_Q7ZYU7 Cirbp-prov protein n=1 Tax=Xenopus laevis RepID... 108 4e-22
UniRef100_Q08529 Glycine-rich protein n=1 Tax=Nicotiana tabacum ... 108 4e-22
UniRef100_B9HL62 Predicted protein n=1 Tax=Populus trichocarpa R... 108 4e-22
UniRef100_B9H173 Predicted protein n=1 Tax=Populus trichocarpa R... 108 4e-22
UniRef100_Q5RCK0 Putative uncharacterized protein DKFZp469I0118 ... 108 4e-22
UniRef100_B4PWI2 GE17772 n=1 Tax=Drosophila yakuba RepID=B4PWI2_... 108 4e-22
UniRef100_B4JMZ8 GH24721 n=1 Tax=Drosophila grimshawi RepID=B4JM... 108 4e-22
UniRef100_O93235 Cold-inducible RNA-binding protein n=1 Tax=Xeno... 108 4e-22
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 107 5e-22
UniRef100_Q6PFA7 Hnrpa3 protein (Fragment) n=1 Tax=Mus musculus ... 107 5e-22
UniRef100_A0K9V3 Putative uncharacterized protein n=1 Tax=Burkho... 107 5e-22
UniRef100_A7QMY0 Chromosome undetermined scaffold_129, whole gen... 107 5e-22
UniRef100_A7PM30 Chromosome chr14 scaffold_21, whole genome shot... 107 5e-22
UniRef100_C0H6E2 Putative cuticle protein n=1 Tax=Bombyx mori Re... 107 5e-22
UniRef100_B4PQR7 GE23791 n=1 Tax=Drosophila yakuba RepID=B4PQR7_... 107 5e-22
UniRef100_B4NC76 GK25807 n=1 Tax=Drosophila willistoni RepID=B4N... 107 5e-22
UniRef100_B4L7M4 GI11200 n=1 Tax=Drosophila mojavensis RepID=B4L... 107 5e-22
UniRef100_B3P5S1 GG11603 n=1 Tax=Drosophila erecta RepID=B3P5S1_... 107 5e-22
UniRef100_B3P425 GG17033 n=1 Tax=Drosophila erecta RepID=B3P425_... 107 5e-22
UniRef100_UPI00006A00C5 UPI00006A00C5 related cluster n=1 Tax=Xe... 107 6e-22
UniRef100_Q6DRJ8 Nucleolin n=1 Tax=Danio rerio RepID=Q6DRJ8_DANRE 107 6e-22
UniRef100_Q28IQ9 Cold inducible RNA binding protein n=1 Tax=Xeno... 107 6e-22
UniRef100_Q9FIQ2 Genomic DNA, chromosome 5, P1 clone:MZA15 n=1 T... 107 6e-22
UniRef100_B9SCP7 Dc50, putative n=1 Tax=Ricinus communis RepID=B... 107 6e-22
UniRef100_B9N506 Predicted protein n=1 Tax=Populus trichocarpa R... 107 6e-22
UniRef100_A9SKS2 Predicted protein n=1 Tax=Physcomitrella patens... 107 6e-22
UniRef100_A8IBU6 Nuclear ribonucleoprotein n=1 Tax=Chlamydomonas... 107 6e-22