[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
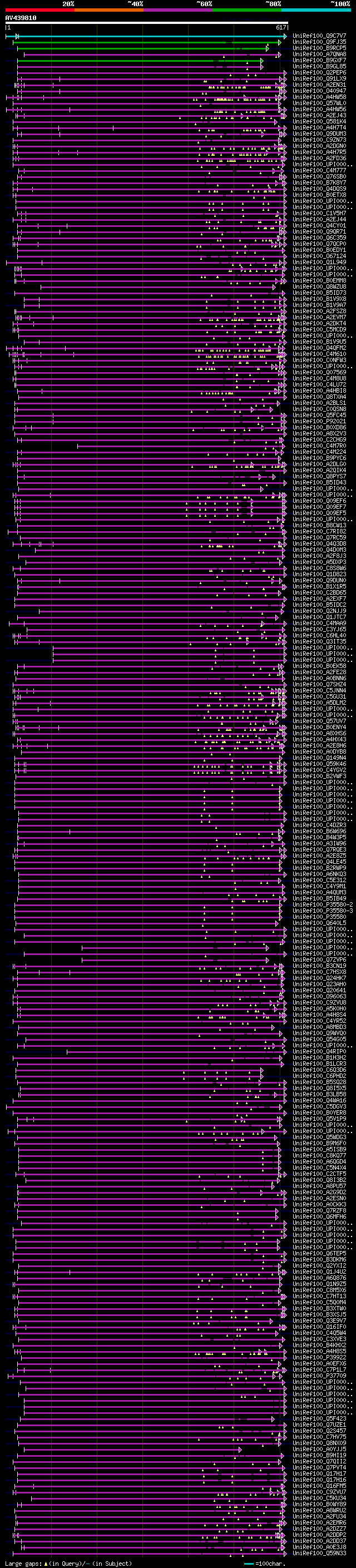
Query= AV439810 APD17e11_f
(617 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q9C7V7 Putative uncharacterized protein F15H21.4 n=1 T... 357 3e-97
UniRef100_Q9FJ35 Myosin heavy chain-like protein n=2 Tax=Arabido... 196 1e-48
UniRef100_B9RCP5 Centromeric protein E, putative n=1 Tax=Ricinus... 121 5e-26
UniRef100_A7QNA8 Chromosome chr2 scaffold_132, whole genome shot... 115 2e-24
UniRef100_B9GXF7 Predicted protein n=1 Tax=Populus trichocarpa R... 115 3e-24
UniRef100_B9GL85 Predicted protein n=1 Tax=Populus trichocarpa R... 111 4e-23
UniRef100_Q2PEP6 Putative myosin heavy chain-like protein n=1 Ta... 108 5e-22
UniRef100_Q91LX9 ORF73 n=1 Tax=Human herpesvirus 8 RepID=Q91LX9_... 81 8e-14
UniRef100_A2EN31 Viral A-type inclusion protein, putative n=1 Ta... 79 3e-13
UniRef100_O40947 ORF 73 n=1 Tax=Human herpesvirus 8 RepID=O40947... 77 9e-13
UniRef100_A4HW58 Kinesin K39, putative n=1 Tax=Leishmania infant... 75 4e-12
UniRef100_Q57WL0 Putative uncharacterized protein n=1 Tax=Trypan... 75 6e-12
UniRef100_A4HW56 Kinesin K39, putative (Fragment) n=1 Tax=Leishm... 75 6e-12
UniRef100_A2EJ43 Viral A-type inclusion protein, putative n=1 Ta... 74 1e-11
UniRef100_Q581K4 Putative uncharacterized protein n=2 Tax=Trypan... 74 1e-11
UniRef100_A4H7T4 Kinesin K39, putative (Fragment) n=1 Tax=Leishm... 74 1e-11
UniRef100_Q9DUM3 Latent nuclear antigen (Fragment) n=1 Tax=Human... 73 2e-11
UniRef100_C9ZN73 Putative uncharacterized protein n=1 Tax=Trypan... 73 2e-11
UniRef100_A2DGN0 Viral A-type inclusion protein, putative n=1 Ta... 72 4e-11
UniRef100_A4H7R5 Kinesin K39, putative n=1 Tax=Leishmania brazil... 72 5e-11
UniRef100_A2FD36 Viral A-type inclusion protein, putative n=1 Ta... 72 5e-11
UniRef100_UPI00006CF26E Viral A-type inclusion protein repeat co... 71 8e-11
UniRef100_C4M777 SMC4 protein, putative n=1 Tax=Entamoeba histol... 71 8e-11
UniRef100_Q76SB0 ORF 73 n=2 Tax=Human herpesvirus 8 RepID=Q76SB0... 70 1e-10
UniRef100_B7K8Y7 BRCT domain protein n=1 Tax=Cyanothece sp. PCC ... 70 1e-10
UniRef100_Q4DQS9 Putative uncharacterized protein (Fragment) n=1... 70 1e-10
UniRef100_B0ETX8 GRIP domain-containing protein RUD3, putative n... 70 1e-10
UniRef100_UPI000175F573 PREDICTED: similar to myosin, heavy poly... 70 2e-10
UniRef100_UPI0001A2D62C UPI0001A2D62C related cluster n=1 Tax=Da... 70 2e-10
UniRef100_C1V5H7 Condensin subunit Smc n=1 Tax=Halogeometricum b... 69 2e-10
UniRef100_A2EJ44 Viral A-type inclusion protein, putative n=1 Ta... 69 3e-10
UniRef100_Q4CY01 Putative uncharacterized protein (Fragment) n=1... 69 4e-10
UniRef100_Q9QR71 ORF73 n=1 Tax=Human herpesvirus 8 type P (isola... 68 5e-10
UniRef100_Q6C359 YALI0F02387p n=1 Tax=Yarrowia lipolytica RepID=... 68 5e-10
UniRef100_Q7QCP0 AGAP002737-PA n=1 Tax=Anopheles gambiae RepID=Q... 68 7e-10
UniRef100_B0EDY1 Tropomyosin isoforms c/e, putative n=1 Tax=Enta... 68 7e-10
UniRef100_O67124 Probable DNA double-strand break repair rad50 A... 68 7e-10
UniRef100_Q1L949 Novel protein (Fragment) n=2 Tax=Danio rerio Re... 67 9e-10
UniRef100_UPI00006CB2DA Viral A-type inclusion protein repeat co... 67 9e-10
UniRef100_UPI00004D9369 Centrosomal protein 2 (Centrosomal Nek2-... 67 9e-10
UniRef100_B0EMM8 Myosin-2 heavy chain, non muscle, putative n=1 ... 67 9e-10
UniRef100_Q8WZU8 Putative uncharacterized protein n=1 Tax=Neuros... 67 9e-10
UniRef100_B5ID73 Putative uncharacterized protein n=1 Tax=Acidul... 67 9e-10
UniRef100_B1V9X8 Putative uncharacterized protein n=1 Tax=Candid... 67 1e-09
UniRef100_B1V9A7 Putative uncharacterized protein n=1 Tax=Candid... 67 1e-09
UniRef100_A2FSZ8 Viral A-type inclusion protein, putative n=1 Ta... 67 1e-09
UniRef100_A2EVM7 DNA-directed RNA polymerase, omega subunit fami... 67 1e-09
UniRef100_A2DKT4 Actinin, putative n=1 Tax=Trichomonas vaginalis... 67 1e-09
UniRef100_C5MCD9 Putative uncharacterized protein n=1 Tax=Candid... 67 1e-09
UniRef100_UPI0001926231 PREDICTED: similar to predicted protein ... 67 2e-09
UniRef100_B1V9U5 Putative uncharacterized protein n=1 Tax=Candid... 67 2e-09
UniRef100_Q4QFM2 Kinesin K39, putative n=1 Tax=Leishmania major ... 67 2e-09
UniRef100_C4M610 Viral A-type inclusion protein repeat, putative... 67 2e-09
UniRef100_C0NFW3 RNA polymerase Rpb1 C-terminal repeat domain-co... 67 2e-09
UniRef100_UPI00006A154D Centrosomal protein 2 (Centrosomal Nek2-... 66 2e-09
UniRef100_Q07569 Myosin heavy chain n=1 Tax=Entamoeba histolytic... 66 2e-09
UniRef100_C4M8U8 Putative uncharacterized protein n=1 Tax=Entamo... 66 2e-09
UniRef100_C4LU72 Myosin heavy chain n=1 Tax=Entamoeba histolytic... 66 2e-09
UniRef100_A4HBI8 Putative uncharacterized protein (Fragment) n=1... 66 2e-09
UniRef100_Q8TXA4 Uncharacterized protein n=1 Tax=Methanopyrus ka... 66 2e-09
UniRef100_A2BLS1 Uncharacterized archaeal coiled-coil protein n=... 66 2e-09
UniRef100_C0QSN8 Chromosome segregation protein SMC n=1 Tax=Pers... 66 3e-09
UniRef100_Q5FC45 Protein T10G3.5b, confirmed by transcript evide... 66 3e-09
UniRef100_P92021 Protein T10G3.5a, confirmed by transcript evide... 66 3e-09
UniRef100_B0XD86 Mushroom body defect protein n=1 Tax=Culex quin... 66 3e-09
UniRef100_A8X2V3 C. briggsae CBR-EEA-1 protein n=1 Tax=Caenorhab... 66 3e-09
UniRef100_C2CHG9 Putative uncharacterized protein n=1 Tax=Anaero... 65 3e-09
UniRef100_C4M7R0 Putative uncharacterized protein n=1 Tax=Entamo... 65 3e-09
UniRef100_C4M224 Villidin, putative n=1 Tax=Entamoeba histolytic... 65 3e-09
UniRef100_B9PYC6 Putative uncharacterized protein n=1 Tax=Toxopl... 65 3e-09
UniRef100_A2DLG0 Viral A-type inclusion protein, putative n=1 Ta... 65 3e-09
UniRef100_A2QIK4 Similarity to microtubule binding protein D-CLI... 65 3e-09
UniRef100_Q8PYS7 Conserved protein n=1 Tax=Methanosarcina mazei ... 65 3e-09
UniRef100_B5ID43 SMC proteins Flexible Hinge Domain n=1 Tax=Acid... 65 3e-09
UniRef100_UPI000180CBAB PREDICTED: similar to M-phase phosphopro... 65 4e-09
UniRef100_UPI000151B79A hypothetical protein PGUG_04173 n=1 Tax=... 65 4e-09
UniRef100_Q09EF6 Abnormal dye filling protein 14, isoform c, par... 65 4e-09
UniRef100_Q09EF7 Abnormal dye filling protein 14, isoform b, par... 65 4e-09
UniRef100_Q09EF5 Abnormal dye filling protein 14, isoform a, par... 65 4e-09
UniRef100_UPI0000E46599 PREDICTED: hypothetical protein, partial... 65 4e-09
UniRef100_B8CW13 Chromosome segregation protein SMC n=1 Tax=Halo... 65 4e-09
UniRef100_C7RI82 Copper amine oxidase domain protein n=1 Tax=Ana... 65 4e-09
UniRef100_Q7RC59 Putative uncharacterized protein PY05925 (Fragm... 65 4e-09
UniRef100_Q4Q3D8 Putative uncharacterized protein n=1 Tax=Leishm... 65 4e-09
UniRef100_Q4D0M3 Putative uncharacterized protein n=1 Tax=Trypan... 65 4e-09
UniRef100_A2F8J3 Kinetoplast-associated protein, putative n=1 Ta... 65 4e-09
UniRef100_A5DXP3 Putative uncharacterized protein n=1 Tax=Lodder... 65 4e-09
UniRef100_C8S8W6 Chromosome segregation protein SMC n=1 Tax=Ferr... 65 4e-09
UniRef100_Q1D823 Adventurous-gliding motility protein Z n=1 Tax=... 65 4e-09
UniRef100_Q9DUN0 Orf73 n=1 Tax=Human herpesvirus 8 RepID=Q9DUN0_... 65 6e-09
UniRef100_B1X1R5 Putative uncharacterized protein n=1 Tax=Cyanot... 65 6e-09
UniRef100_C2BD65 Viral A family inclusion protein (Fragment) n=1... 65 6e-09
UniRef100_A2EXF7 Putative uncharacterized protein n=1 Tax=Tricho... 65 6e-09
UniRef100_B5IDC2 Putative uncharacterized protein n=1 Tax=Acidul... 65 6e-09
UniRef100_Q2NJJ9 Putative uncharacterized protein n=1 Tax=Aster ... 64 8e-09
UniRef100_Q1JTC7 Putative uncharacterized protein n=1 Tax=Toxopl... 64 8e-09
UniRef100_C4MAA9 Putative uncharacterized protein n=1 Tax=Entamo... 64 8e-09
UniRef100_C3YJ65 Putative uncharacterized protein n=1 Tax=Branch... 64 8e-09
UniRef100_C6HL40 RNA polymerase Rpb1 C-terminal repeat domain-co... 64 8e-09
UniRef100_Q3IT35 Chromosome partition protein n=1 Tax=Natronomon... 64 8e-09
UniRef100_UPI0000DA32F1 PREDICTED: similar to ciliary rootlet co... 64 1e-08
UniRef100_UPI0000DA3207 PREDICTED: similar to ciliary rootlet co... 64 1e-08
UniRef100_UPI0001B7A7E1 UPI0001B7A7E1 related cluster n=1 Tax=Ra... 64 1e-08
UniRef100_B0EK58 Villin, putative n=1 Tax=Entamoeba dispar SAW76... 64 1e-08
UniRef100_A2FE28 Putative uncharacterized protein n=1 Tax=Tricho... 64 1e-08
UniRef100_A0BNN6 Chromosome undetermined scaffold_119, whole gen... 64 1e-08
UniRef100_Q7SHZ4 Predicted protein n=1 Tax=Neurospora crassa Rep... 64 1e-08
UniRef100_C5JNN4 RNA polymerase Rpb1 C-terminal repeat domain-co... 64 1e-08
UniRef100_C5GU31 RNA polymerase Rpb1 C-terminal repeat domain-co... 64 1e-08
UniRef100_A5DLM2 Putative uncharacterized protein n=1 Tax=Pichia... 64 1e-08
UniRef100_UPI00003C004D PREDICTED: similar to centromere protein... 64 1e-08
UniRef100_UPI000012310B Hypothetical protein CBG13429 n=1 Tax=Ca... 64 1e-08
UniRef100_Q57UV7 Kinesin, putative n=1 Tax=Trypanosoma brucei Re... 64 1e-08
UniRef100_B0ENY4 Intracellular protein transport protein USO1, p... 64 1e-08
UniRef100_A8XHS6 C. briggsae CBR-DYF-14 protein n=1 Tax=Caenorha... 64 1e-08
UniRef100_A4HX43 Kinesin, putative (Fragment) n=1 Tax=Leishmania... 64 1e-08
UniRef100_A2E8H6 Viral A-type inclusion protein, putative n=1 Ta... 64 1e-08
UniRef100_A0DYB8 Chromosome undetermined scaffold_7, whole genom... 64 1e-08
UniRef100_Q149N4 Myosin, heavy chain 10, non-muscle n=1 Tax=Homo... 64 1e-08
UniRef100_Q59K46 Likely vesicular transport factor Uso1p (Fragme... 64 1e-08
UniRef100_C4YGV2 Putative uncharacterized protein n=1 Tax=Candid... 64 1e-08
UniRef100_B2VWF3 Intracellular protein transport protein (UsoA) ... 64 1e-08
UniRef100_UPI000186DD89 retiin, putative n=1 Tax=Pediculus human... 63 2e-08
UniRef100_UPI0000E2492B PREDICTED: myosin, heavy polypeptide 10,... 63 2e-08
UniRef100_UPI0000E2492A PREDICTED: myosin, heavy polypeptide 10,... 63 2e-08
UniRef100_UPI0000E24929 PREDICTED: myosin, heavy polypeptide 10,... 63 2e-08
UniRef100_UPI0000E24928 PREDICTED: myosin, heavy polypeptide 10,... 63 2e-08
UniRef100_UPI0000E1E884 PREDICTED: WD repeat domain 65 n=1 Tax=P... 63 2e-08
UniRef100_UPI0000D998D4 PREDICTED: similar to CG4329-PA, isoform... 63 2e-08
UniRef100_C4DZR3 Putative uncharacterized protein (Fragment) n=1... 63 2e-08
UniRef100_B6W696 Putative uncharacterized protein n=1 Tax=Anaero... 63 2e-08
UniRef100_B4W3P5 Tetratricopeptide repeat domain protein n=1 Tax... 63 2e-08
UniRef100_A3IW96 DNA ligase n=1 Tax=Cyanothece sp. CCY0110 RepID... 63 2e-08
UniRef100_Q7RQE3 Putative uncharacterized protein PY01156 n=1 Ta... 63 2e-08
UniRef100_A2E8Z5 Viral A-type inclusion protein, putative n=1 Ta... 63 2e-08
UniRef100_Q4LE45 MYH10 variant protein (Fragment) n=2 Tax=Homini... 63 2e-08
UniRef100_B2RWP9 MYH10 protein n=1 Tax=Homo sapiens RepID=B2RWP9... 63 2e-08
UniRef100_A6NKQ3 Uncharacterized protein ENSP00000361571 n=2 Tax... 63 2e-08
UniRef100_C5E312 KLTH0H09438p n=1 Tax=Lachancea thermotolerans C... 63 2e-08
UniRef100_C4Y9M1 Putative uncharacterized protein n=1 Tax=Clavis... 63 2e-08
UniRef100_A4QUM3 Predicted protein n=1 Tax=Magnaporthe grisea Re... 63 2e-08
UniRef100_B5IB49 SMC proteins Flexible Hinge Domain n=1 Tax=Acid... 63 2e-08
UniRef100_P35580-2 Isoform 2 of Myosin-10 n=1 Tax=Homo sapiens R... 63 2e-08
UniRef100_P35580-3 Isoform 3 of Myosin-10 n=1 Tax=Homo sapiens R... 63 2e-08
UniRef100_P35580 Myosin-10 n=1 Tax=Homo sapiens RepID=MYH10_HUMAN 63 2e-08
UniRef100_Q640L5 Coiled-coil domain-containing protein 18 n=1 Ta... 63 2e-08
UniRef100_UPI0000E46049 PREDICTED: similar to myosin, heavy poly... 63 2e-08
UniRef100_UPI00005A4998 PREDICTED: similar to Huntingtin interac... 63 2e-08
UniRef100_UPI0000122375 Hypothetical protein CBG02025 n=1 Tax=Ca... 63 2e-08
UniRef100_UPI0000568DAE sarcolemma associated protein n=1 Tax=Da... 63 2e-08
UniRef100_UPI0000EB05FB Huntingtin-interacting protein 1-related... 63 2e-08
UniRef100_Q7ZVP6 Sarcolemma associated protein n=1 Tax=Danio rer... 63 2e-08
UniRef100_B3CN19 Putative uncharacterized protein n=1 Tax=Wolbac... 63 2e-08
UniRef100_C7HSX8 Surface protein n=1 Tax=Anaerococcus vaginalis ... 63 2e-08
UniRef100_Q24HK7 Viral A-type inclusion protein repeat containin... 63 2e-08
UniRef100_Q23AH0 Putative uncharacterized protein n=1 Tax=Tetrah... 63 2e-08
UniRef100_Q20641 Non-muscle myosin protein 1 n=1 Tax=Caenorhabdi... 63 2e-08
UniRef100_O96063 Myosin heavy chain (Fragment) n=1 Tax=Dugesia j... 63 2e-08
UniRef100_C9ZVU8 Kinesin putative, (Fragment) n=1 Tax=Trypanosom... 63 2e-08
UniRef100_A5K0H0 Putative uncharacterized protein n=1 Tax=Plasmo... 63 2e-08
UniRef100_A4H8S4 Kinesin, putative n=1 Tax=Leishmania braziliens... 63 2e-08
UniRef100_C4YR52 Putative uncharacterized protein n=1 Tax=Candid... 63 2e-08
UniRef100_A8MBD3 Chromosome segregation ATPase-like protein n=1 ... 63 2e-08
UniRef100_Q9WVQ0 Polyamine-modulated factor 1-binding protein 1 ... 63 2e-08
UniRef100_Q54G05 Putative leucine-rich repeat-containing protein... 63 2e-08
UniRef100_UPI00016E2754 UPI00016E2754 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_Q4RIP0 Chromosome 7 SCAF15042, whole genome shotgun se... 62 3e-08
UniRef100_B1H3H2 LOC100145631 protein n=1 Tax=Xenopus (Silurana)... 62 3e-08
UniRef100_B1LCR3 Chromosome segregation protein SMC n=2 Tax=Ther... 62 3e-08
UniRef100_C6Q3D6 SMC domain protein n=1 Tax=Thermoanaerobacter m... 62 3e-08
UniRef100_C6PHD2 SMC domain protein n=1 Tax=Thermoanaerobacter i... 62 3e-08
UniRef100_B5SQ28 Putative surface protein n=1 Tax=Streptococcus ... 62 3e-08
UniRef100_Q8I5X5 Conserved Plasmodium protein n=1 Tax=Plasmodium... 62 3e-08
UniRef100_B3LB58 Putative uncharacterized protein n=1 Tax=Plasmo... 62 3e-08
UniRef100_Q4WA16 Vesicle-mediated transport protein (Imh1), puta... 62 3e-08
UniRef100_C5DGV3 KLTH0D08558p n=1 Tax=Lachancea thermotolerans C... 62 3e-08
UniRef100_B0YER8 Vesicle-mediated transport protein (Imh1), puta... 62 3e-08
UniRef100_Q5V1P9 Chromosome segregation protein n=1 Tax=Haloarcu... 62 3e-08
UniRef100_UPI0001758011 PREDICTED: similar to hyaluronan-mediate... 62 4e-08
UniRef100_UPI0000F2C3AC PREDICTED: similar to basalin n=1 Tax=Mo... 62 4e-08
UniRef100_Q5WDG3 Metalloendopeptidase n=1 Tax=Bacillus clausii K... 62 4e-08
UniRef100_B9M6F0 Chromosome segregation protein SMC n=1 Tax=Geob... 62 4e-08
UniRef100_A5ISB9 Condensin subunit Smc n=12 Tax=Staphylococcus a... 62 4e-08
UniRef100_C8KQ77 Chromosome segregation SMC protein n=1 Tax=Stap... 62 4e-08
UniRef100_A6QGD4 Chromosome segregation SMC protein n=7 Tax=Stap... 62 4e-08
UniRef100_C5N4X4 SMC superfamily ATP-binding chromosome segregat... 62 4e-08
UniRef100_C2CTF5 Putative uncharacterized protein n=1 Tax=Gardne... 62 4e-08
UniRef100_Q8I3B2 Putative uncharacterized protein n=1 Tax=Plasmo... 62 4e-08
UniRef100_A8PU57 Putative uncharacterized protein n=1 Tax=Brugia... 62 4e-08
UniRef100_A2G9D2 Putative uncharacterized protein n=1 Tax=Tricho... 62 4e-08
UniRef100_A2ESN0 Viral A-type inclusion protein, putative n=1 Ta... 62 4e-08
UniRef100_A0CKK3 Chromosome undetermined scaffold_2, whole genom... 62 4e-08
UniRef100_Q7RZF8 Predicted protein n=1 Tax=Neurospora crassa Rep... 62 4e-08
UniRef100_Q6MFH6 Related to nucleoprotein TPR n=1 Tax=Neurospora... 62 4e-08
UniRef100_UPI000192558E PREDICTED: similar to myosin heavy chain... 62 5e-08
UniRef100_UPI0001A2D16E hyaluronan mediated motility receptor n=... 62 5e-08
UniRef100_UPI0001A2D16D hyaluronan mediated motility receptor n=... 62 5e-08
UniRef100_UPI00016E0862 UPI00016E0862 related cluster n=1 Tax=Ta... 62 5e-08
UniRef100_UPI00016E0844 UPI00016E0844 related cluster n=1 Tax=Ta... 62 5e-08
UniRef100_Q6TEP5 Hyaluronan-mediated motility receptor n=1 Tax=D... 62 5e-08
UniRef100_B3DKM6 Hmmr protein n=1 Tax=Danio rerio RepID=B3DKM6_D... 62 5e-08
UniRef100_Q2YXI2 Chromosome segregation SMC protein n=1 Tax=Stap... 62 5e-08
UniRef100_Q1J4U2 Putative surface protein n=1 Tax=Streptococcus ... 62 5e-08
UniRef100_A6Q876 DNA double-strand break repair protein n=1 Tax=... 62 5e-08
UniRef100_Q1N9Z5 Putative uncharacterized protein n=1 Tax=Sphing... 62 5e-08
UniRef100_C8M5X6 Condensin subunit Smc n=1 Tax=Staphylococcus au... 62 5e-08
UniRef100_C7HT13 Surface protein n=1 Tax=Anaerococcus vaginalis ... 62 5e-08
UniRef100_C5Q0M4 Chromosome segregation SMC protein n=1 Tax=Stap... 62 5e-08
UniRef100_B3XTW0 Putative surface protein (Fragment) n=1 Tax=Ure... 62 5e-08
UniRef100_B3XSJ5 Putative surface protein n=1 Tax=Ureaplasma ure... 62 5e-08
UniRef100_Q3E9V7 Putative uncharacterized protein At4g27595.1 n=... 62 5e-08
UniRef100_Q16IF0 Condensin, SMC5-subunit, putative (Fragment) n=... 62 5e-08
UniRef100_C4Q5W4 Tropomyosin, putative n=1 Tax=Schistosoma manso... 62 5e-08
UniRef100_C3XVE3 Putative uncharacterized protein n=1 Tax=Branch... 62 5e-08
UniRef100_B4KHX2 GI14952 n=1 Tax=Drosophila mojavensis RepID=B4K... 62 5e-08
UniRef100_A4H8S5 Kinesin, putative (Fragment) n=1 Tax=Leishmania... 62 5e-08
UniRef100_P39922 Myosin heavy chain, clone 203 (Fragment) n=2 Ta... 62 5e-08
UniRef100_A0EFX6 Chromosome undetermined scaffold_94, whole geno... 62 5e-08
UniRef100_C7P1L7 Chromosome segregation protein SMC n=1 Tax=Halo... 62 5e-08
UniRef100_P37709 Trichohyalin n=1 Tax=Oryctolagus cuniculus RepI... 62 5e-08
UniRef100_UPI0000E48979 PREDICTED: similar to kinesin-related pr... 61 6e-08
UniRef100_UPI0000D55EA0 PREDICTED: similar to viral A-type inclu... 61 6e-08
UniRef100_UPI00015A4D2D UPI00015A4D2D related cluster n=1 Tax=Da... 61 6e-08
UniRef100_UPI00016E64AD UPI00016E64AD related cluster n=1 Tax=Ta... 61 6e-08
UniRef100_UPI00016E64AC UPI00016E64AC related cluster n=1 Tax=Ta... 61 6e-08
UniRef100_UPI00016E648C UPI00016E648C related cluster n=1 Tax=Ta... 61 6e-08
UniRef100_Q5F423 Putative uncharacterized protein n=1 Tax=Gallus... 61 6e-08
UniRef100_Q7UZE1 Similar to myosin heavy chain n=1 Tax=Rhodopire... 61 6e-08
UniRef100_Q2S457 Chromosome segregation protein SMC n=1 Tax=Sali... 61 6e-08
UniRef100_C7HV75 Putative uncharacterized protein n=1 Tax=Anaero... 61 6e-08
UniRef100_Q8NX09 Chromosome segregation SMC protein n=3 Tax=Stap... 61 6e-08
UniRef100_A0YJJ5 Putative uncharacterized protein n=1 Tax=Lyngby... 61 6e-08
UniRef100_B9HI19 Predicted protein n=1 Tax=Populus trichocarpa R... 61 6e-08
UniRef100_Q7QII2 AGAP006884-PA n=1 Tax=Anopheles gambiae RepID=Q... 61 6e-08
UniRef100_Q7PVT4 AGAP009189-PA (Fragment) n=1 Tax=Anopheles gamb... 61 6e-08
UniRef100_Q17H17 Slender lobes, putative n=1 Tax=Aedes aegypti R... 61 6e-08
UniRef100_Q17H16 Slender lobes, putative n=1 Tax=Aedes aegypti R... 61 6e-08
UniRef100_Q16FM5 LL5 beta protein, putative n=1 Tax=Aedes aegypt... 61 6e-08
UniRef100_C9ZVU7 Kinesin, putative, (Fragment) n=1 Tax=Trypanoso... 61 6e-08
UniRef100_C5KU34 Putative uncharacterized protein n=1 Tax=Perkin... 61 6e-08
UniRef100_B0WY89 Microtubule binding protein D-CLIP-190 n=1 Tax=... 61 6e-08
UniRef100_A8WRU2 C. briggsae CBR-NMY-1 protein n=1 Tax=Caenorhab... 61 6e-08
UniRef100_A2FU34 Putative uncharacterized protein n=1 Tax=Tricho... 61 6e-08
UniRef100_A2EMR6 Viral A-type inclusion protein, putative n=1 Ta... 61 6e-08
UniRef100_A2DZZ7 Smooth muscle caldesmon, putative n=1 Tax=Trich... 61 6e-08
UniRef100_A2DDP2 Viral A-type inclusion protein, putative n=1 Ta... 61 6e-08
UniRef100_A2DD37 Viral A-type inclusion protein, putative n=1 Ta... 61 6e-08
UniRef100_A0E3J8 Chromosome undetermined scaffold_76, whole geno... 61 6e-08
UniRef100_Q59N33 Putative uncharacterized protein MYO1 n=1 Tax=C... 61 6e-08
UniRef100_C5DFC9 KLTH0D14102p n=1 Tax=Lachancea thermotolerans C... 61 6e-08
UniRef100_A1DB41 Putative uncharacterized protein n=1 Tax=Neosar... 61 6e-08
UniRef100_A5YS38 Chromosome segregation protein n=1 Tax=uncultur... 61 6e-08
UniRef100_UPI0000D9C6EF PREDICTED: similar to centrosomal protei... 61 8e-08
UniRef100_UPI0000D9C6EE PREDICTED: similar to centrosomal protei... 61 8e-08
UniRef100_UPI00006CE95F Viral A-type inclusion protein repeat co... 61 8e-08
UniRef100_UPI00005C20FD kinesin family member 3B n=2 Tax=Bos tau... 61 8e-08
UniRef100_Q4RQT6 Chromosome 2 SCAF15004, whole genome shotgun se... 61 8e-08
UniRef100_B0S1Y6 Putative chimeric erythrocyte-binding protein n... 61 8e-08
UniRef100_Q4C442 BRCT n=1 Tax=Crocosphaera watsonii WH 8501 RepI... 61 8e-08
UniRef100_B6Y794 Putative uncharacterized protein n=1 Tax=Wolbac... 61 8e-08
UniRef100_B5W3Z8 Methyltransferase FkbM family n=1 Tax=Arthrospi... 61 8e-08
UniRef100_Q4DSZ8 Putative uncharacterized protein n=1 Tax=Trypan... 61 8e-08
UniRef100_Q4DRJ0 Putative uncharacterized protein n=1 Tax=Trypan... 61 8e-08
UniRef100_Q4DG71 Putative uncharacterized protein (Fragment) n=1... 61 8e-08
UniRef100_Q16XH2 RHC18, putative n=1 Tax=Aedes aegypti RepID=Q16... 61 8e-08
UniRef100_C4Q5W7 Tropomyosin, putative n=1 Tax=Schistosoma manso... 61 8e-08
UniRef100_B9PJI1 Structural maintenance of chromosomes 6 smc6, p... 61 8e-08
UniRef100_B4J2R7 GH16034 n=1 Tax=Drosophila grimshawi RepID=B4J2... 61 8e-08
UniRef100_B0EUK2 Putative uncharacterized protein n=1 Tax=Entamo... 61 8e-08
UniRef100_A7RUF8 Predicted protein (Fragment) n=1 Tax=Nematostel... 61 8e-08
UniRef100_A2GM00 Putative uncharacterized protein (Fragment) n=1... 61 8e-08
UniRef100_A2EYA1 Viral A-type inclusion protein, putative n=1 Ta... 61 8e-08
UniRef100_A2DNX6 Viral A-type inclusion protein, putative n=1 Ta... 61 8e-08
UniRef100_A5DD85 Putative uncharacterized protein n=1 Tax=Pichia... 61 8e-08
UniRef100_UPI000180C7F9 PREDICTED: similar to myomegalin n=1 Tax... 60 1e-07
UniRef100_UPI000155F65F PREDICTED: similar to RUN and FYVE domai... 60 1e-07
UniRef100_UPI000151ACE1 hypothetical protein PGUG_01236 n=1 Tax=... 60 1e-07
UniRef100_UPI0000F2E4F7 PREDICTED: similar to GTPase, IMAP famil... 60 1e-07
UniRef100_UPI0001A2C816 UPI0001A2C816 related cluster n=1 Tax=Da... 60 1e-07
UniRef100_UPI000060F511 myosin, heavy polypeptide 10, non-muscle... 60 1e-07
UniRef100_UPI000060F510 myosin, heavy polypeptide 10, non-muscle... 60 1e-07
UniRef100_Q98UK0 Lamin B2 n=1 Tax=Carassius auratus RepID=Q98UK0... 60 1e-07
UniRef100_Q801N8 LOC398577 protein (Fragment) n=1 Tax=Xenopus la... 60 1e-07
UniRef100_Q789A6 Nonmuscle myosin heavy chain n=2 Tax=Gallus gal... 60 1e-07
UniRef100_Q789A5 Nonmuscle myosin heavy chain n=1 Tax=Gallus gal... 60 1e-07
UniRef100_Q789A4 Nonmuscle myosin heavy chain n=1 Tax=Gallus gal... 60 1e-07
UniRef100_Q02015 Nonmuscle myosin heavy chain n=2 Tax=Gallus gal... 60 1e-07
UniRef100_Q8I951 SMC4 protein n=1 Tax=Anopheles gambiae RepID=Q8... 60 1e-07
UniRef100_Q7Q3D7 AGAP007826-PA n=1 Tax=Anopheles gambiae RepID=Q... 60 1e-07
UniRef100_C4Q358 Tropomyosin, putative n=1 Tax=Schistosoma manso... 60 1e-07
UniRef100_C3ZL86 Putative uncharacterized protein n=1 Tax=Branch... 60 1e-07
UniRef100_B6KA29 Chromosome segregation protein, putative n=2 Ta... 60 1e-07
UniRef100_B4I434 GM10613 n=1 Tax=Drosophila sechellia RepID=B4I4... 60 1e-07
UniRef100_B3L3Y4 Putative uncharacterized protein n=1 Tax=Plasmo... 60 1e-07
UniRef100_A2FP55 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A2EZE6 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A2ETW9 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A2ERL6 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A2DLG1 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A0CPG2 Chromosome undetermined scaffold_23, whole geno... 60 1e-07
UniRef100_C4XYQ3 Putative uncharacterized protein n=1 Tax=Clavis... 60 1e-07
UniRef100_A7TQ63 Putative uncharacterized protein n=1 Tax=Vander... 60 1e-07
UniRef100_A7F074 Putative uncharacterized protein n=1 Tax=Sclero... 60 1e-07
UniRef100_Q96YR5 DNA double-strand break repair rad50 ATPase n=1... 60 1e-07
UniRef100_UPI00018650AF hypothetical protein BRAFLDRAFT_87034 n=... 60 1e-07
UniRef100_UPI00017F0A51 PREDICTED: similar to KIAA0655 protein n... 60 1e-07
UniRef100_UPI00017F08EE PREDICTED: similar to KIAA0655 protein n... 60 1e-07
UniRef100_UPI00015B5D72 PREDICTED: similar to viral A-type inclu... 60 1e-07
UniRef100_UPI00006CB6F1 hypothetical protein TTHERM_00494240 n=1... 60 1e-07
UniRef100_UPI000023D00A hypothetical protein FG01414.1 n=1 Tax=G... 60 1e-07
UniRef100_B4F6N3 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_B3DL66 Ktn1 protein n=1 Tax=Xenopus (Silurana) tropica... 60 1e-07
UniRef100_C8MEX3 Chromosome segregation protein SMC n=1 Tax=Stap... 60 1e-07
UniRef100_Q7XN10 OSJNBa0008M17.5 protein n=1 Tax=Oryza sativa Ja... 60 1e-07
UniRef100_Q01K10 OSIGBa0126B18.8 protein n=1 Tax=Oryza sativa Re... 60 1e-07
UniRef100_B9FCH2 Putative uncharacterized protein n=1 Tax=Oryza ... 60 1e-07
UniRef100_B8AU60 Putative uncharacterized protein n=1 Tax=Oryza ... 60 1e-07
UniRef100_B0CM79 Centrosomal protein 250kDa, isoform 1 (Predicte... 60 1e-07
UniRef100_Q4QEL9 Kinesin, putative n=1 Tax=Leishmania major RepI... 60 1e-07
UniRef100_Q1ZXP5 Putative uncharacterized protein vilC n=1 Tax=D... 60 1e-07
UniRef100_B4L2T6 GI15438 n=1 Tax=Drosophila mojavensis RepID=B4L... 60 1e-07
UniRef100_B2ZTQ6 Muscle myosin heavy chain n=1 Tax=Sepia esculen... 60 1e-07
UniRef100_A2FQ08 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A2DUI3 Viral A-type inclusion protein, putative n=1 Ta... 60 1e-07
UniRef100_A0EAY8 Chromosome undetermined scaffold_87, whole geno... 60 1e-07
UniRef100_Q4PGJ7 Putative uncharacterized protein n=1 Tax=Ustila... 60 1e-07
UniRef100_Q1DLC4 Putative uncharacterized protein n=1 Tax=Coccid... 60 1e-07
UniRef100_C5P4B0 Viral A-type inclusion protein repeat containin... 60 1e-07
UniRef100_B6QU61 Vesicle-mediated transport protein (Imh1), puta... 60 1e-07
UniRef100_B6K1Y4 Predicted protein n=1 Tax=Schizosaccharomyces j... 60 1e-07
UniRef100_A4R727 Putative uncharacterized protein n=1 Tax=Magnap... 60 1e-07
UniRef100_Q9PTD7 Cingulin n=1 Tax=Xenopus laevis RepID=CING_XENLA 60 1e-07
UniRef100_UPI000156005D PREDICTED: similar to Kinesin-like prote... 60 2e-07
UniRef100_UPI0000E48EEB PREDICTED: similar to Viral A-type inclu... 60 2e-07
UniRef100_UPI0000D9CF94 PREDICTED: similar to Golgi autoantigen,... 60 2e-07
UniRef100_UPI00006CCCFD hypothetical protein TTHERM_00476520 n=1... 60 2e-07
UniRef100_UPI00005A4464 PREDICTED: similar to Kinesin-like prote... 60 2e-07
UniRef100_UPI00017B528F UPI00017B528F related cluster n=1 Tax=Te... 60 2e-07
UniRef100_UPI00017B2B3B UPI00017B2B3B related cluster n=1 Tax=Te... 60 2e-07
UniRef100_UPI000184A0D4 Kinesin-like protein KIF3B (Microtubule ... 60 2e-07
UniRef100_Q4T443 Chromosome undetermined SCAF9830, whole genome ... 60 2e-07
UniRef100_Q4S0W3 Chromosome undetermined SCAF14776, whole genome... 60 2e-07
UniRef100_B0JZG9 LOC100145206 protein n=1 Tax=Xenopus (Silurana)... 60 2e-07
UniRef100_Q8BNH4 Putative uncharacterized protein n=1 Tax=Mus mu... 60 2e-07
UniRef100_Q61771 Kinesin-like protein KIF3B n=3 Tax=Mus musculus... 60 2e-07
UniRef100_Q3UFZ8 Putative uncharacterized protein n=1 Tax=Mus mu... 60 2e-07
UniRef100_Q8RCY8 ATPase involved in DNA repair n=1 Tax=Thermoana... 60 2e-07
UniRef100_B5YIF1 Chromosome segregation SMC protein, putative n=... 60 2e-07
UniRef100_A9BEV5 Exonuclease sbcC n=1 Tax=Petrotoga mobilis SJ95... 60 2e-07
UniRef100_C3WDH6 Predicted protein n=1 Tax=Fusobacterium mortife... 60 2e-07
UniRef100_B7R6Z1 RecF/RecN/SMC N terminal domain, putative n=1 T... 60 2e-07
UniRef100_Q23D90 Putative uncharacterized protein n=1 Tax=Tetrah... 60 2e-07
UniRef100_Q233E2 Putative uncharacterized protein n=1 Tax=Tetrah... 60 2e-07
UniRef100_Q22257 Protein T06E4.1, partially confirmed by transcr... 60 2e-07
UniRef100_B4LUF6 GJ24094 n=1 Tax=Drosophila virilis RepID=B4LUF6... 60 2e-07
UniRef100_A0EHR1 Chromosome undetermined scaffold_97, whole geno... 60 2e-07
UniRef100_Q0U842 Putative uncharacterized protein n=1 Tax=Phaeos... 60 2e-07
UniRef100_Q9V1R8 Smc1 chromosome segregation protein n=1 Tax=Pyr... 60 2e-07
UniRef100_C7P316 Putative uncharacterized protein n=1 Tax=Halomi... 60 2e-07
UniRef100_C5U6T2 Chromosome segregation protein SMC n=1 Tax=Meth... 60 2e-07
UniRef100_Q9GZ69 Tropomyosin n=1 Tax=Mimachlamys nobilis RepID=T... 60 2e-07
UniRef100_O15066 Kinesin-like protein KIF3B n=1 Tax=Homo sapiens... 60 2e-07
UniRef100_UPI0001760282 PREDICTED: im:7147268, partial n=1 Tax=D... 59 2e-07
UniRef100_UPI000175FC42 PREDICTED: similar to centrosomal protei... 59 2e-07
UniRef100_UPI0000DB7C7C PREDICTED: similar to zipper CG15792-PD,... 59 2e-07
UniRef100_UPI0000DA3AD8 PREDICTED: similar to sarcoma antigen NY... 59 2e-07
UniRef100_UPI0000DA3516 PREDICTED: similar to Myosin-11 (Myosin ... 59 2e-07
UniRef100_UPI0000DA3515 PREDICTED: similar to myosin, heavy poly... 59 2e-07
UniRef100_UPI0000DA3514 PREDICTED: similar to Myosin-11 (Myosin ... 59 2e-07
UniRef100_UPI0000DA3513 PREDICTED: similar to Myosin-11 (Myosin ... 59 2e-07
UniRef100_UPI0000DA3311 PREDICTED: similar to Myosin-11 (Myosin ... 59 2e-07
UniRef100_B8ZZW2 Putative uncharacterized protein GCC2 n=2 Tax=H... 59 2e-07
UniRef100_UPI0001A2BB29 UPI0001A2BB29 related cluster n=1 Tax=Da... 59 2e-07
UniRef100_UPI00004D936A Centrosomal protein 2 (Centrosomal Nek2-... 59 2e-07
UniRef100_UPI0001B7B55C similar to CG4329-PA, isoform A (LOC6796... 59 2e-07
UniRef100_UPI0001B7A5F7 UPI0001B7A5F7 related cluster n=1 Tax=Ra... 59 2e-07
UniRef100_UPI0001B7A1F7 Myosin-11 (Myosin heavy chain 11) (Myosi... 59 2e-07
UniRef100_UPI0001B7A19D Myosin-11 (Myosin heavy chain 11) (Myosi... 59 2e-07
UniRef100_UPI0001B7A19C Myosin-11 (Myosin heavy chain 11) (Myosi... 59 2e-07
UniRef100_UPI0001B7A19B Myosin-11 (Myosin heavy chain 11) (Myosi... 59 2e-07
UniRef100_UPI0001B79B84 polyamine modulated factor 1 binding pro... 59 2e-07
UniRef100_UPI0001B79B83 polyamine modulated factor 1 binding pro... 59 2e-07
UniRef100_UPI0000DA247A kinesin family member 3B n=1 Tax=Rattus ... 59 2e-07
UniRef100_UPI0000EB2D48 UPI0000EB2D48 related cluster n=1 Tax=Ca... 59 2e-07
UniRef100_Q6PB65 Myh10 protein (Fragment) n=1 Tax=Mus musculus R... 59 2e-07
UniRef100_Q5SV64 Myosin, heavy polypeptide 10, non-muscle n=1 Ta... 59 2e-07
UniRef100_Q4KMN4 Myh10 protein (Fragment) n=1 Tax=Mus musculus R... 59 2e-07
UniRef100_Q61879 Myosin-10 n=3 Tax=Mus musculus RepID=MYH10_MOUSE 59 2e-07
UniRef100_Q5X238 Effector protein B, substrate of the Dot/Icm se... 59 2e-07
UniRef100_C5CIS4 Chromosome segregation protein SMC n=1 Tax=Kosm... 59 2e-07
UniRef100_B9K9D7 Condensin subunit Smc n=1 Tax=Thermotoga neapol... 59 2e-07
UniRef100_A6V5T2 TnpT protein n=1 Tax=Pseudomonas aeruginosa PA7... 59 2e-07
UniRef100_C2CJ43 Possible surface protein (Fragment) n=1 Tax=Ana... 59 2e-07
UniRef100_Q00VG0 Homology to unknown gene n=1 Tax=Ostreococcus t... 59 2e-07
UniRef100_C1MVG6 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 2e-07
UniRef100_B9SFG7 ATP binding protein, putative n=1 Tax=Ricinus c... 59 2e-07
UniRef100_B9S6T3 GTP binding protein, putative n=1 Tax=Ricinus c... 59 2e-07
UniRef100_Q6X276 Huntingtin interacting protein 1-related n=1 Ta... 59 2e-07
UniRef100_Q9U0S6 Pedal retractor muscle myosin heavy chain (Frag... 59 2e-07
UniRef100_Q7YWF2 Normocyte binding protein 2a n=1 Tax=Plasmodium... 59 2e-07
UniRef100_Q7RGY2 Repeat organellar protein-related (Fragment) n=... 59 2e-07
UniRef100_Q57U93 Putative uncharacterized protein n=1 Tax=Trypan... 59 2e-07
UniRef100_C9ZPQ1 Putative uncharacterized protein n=1 Tax=Trypan... 59 2e-07
UniRef100_C3YUC1 Putative uncharacterized protein n=1 Tax=Branch... 59 2e-07
UniRef100_B9QNS2 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B9QIT4 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B9Q0Z1 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B9PXC5 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B6KNE3 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B6KAK5 Putative uncharacterized protein n=1 Tax=Toxopl... 59 2e-07
UniRef100_B3NPC8 GG20041 n=1 Tax=Drosophila erecta RepID=B3NPC8_... 59 2e-07
UniRef100_B2ZTQ5 Muscle myosin heavy chain n=1 Tax=Loligo bleeke... 59 2e-07
UniRef100_B0WD78 Condensin, SMC5-subunit n=1 Tax=Culex quinquefa... 59 2e-07
UniRef100_A7AMI4 Putative uncharacterized protein n=1 Tax=Babesi... 59 2e-07
UniRef100_A2FKS2 Putative uncharacterized protein n=1 Tax=Tricho... 59 2e-07
UniRef100_A8MT32 Putative uncharacterized protein GCC2 (Fragment... 59 2e-07
UniRef100_Q2UQD3 Dystonin n=1 Tax=Aspergillus oryzae RepID=Q2UQD... 59 2e-07
UniRef100_B9WD59 Uncharacterized protein ymr031c homologue, puta... 59 2e-07
UniRef100_Q9Z221 Polyamine-modulated factor 1-binding protein 1 ... 59 2e-07
UniRef100_Q63862-2 Isoform 2 of Myosin-11 n=1 Tax=Rattus norvegi... 59 2e-07
UniRef100_Q63862-3 Isoform 3 of Myosin-11 n=1 Tax=Rattus norvegi... 59 2e-07
UniRef100_Q63862 Myosin-11 (Fragments) n=1 Tax=Rattus norvegicus... 59 2e-07
UniRef100_Q8IWJ2 GRIP and coiled-coil domain-containing protein ... 59 2e-07
UniRef100_UPI000194DA0D PREDICTED: similar to mCG5221, partial n... 59 3e-07
UniRef100_UPI00017961B8 PREDICTED: coiled-coil domain containing... 59 3e-07
UniRef100_UPI0001760070 PREDICTED: hypothetical protein, partial... 59 3e-07
UniRef100_UPI000155C219 PREDICTED: similar to smooth muscle myos... 59 3e-07
UniRef100_UPI0001552C3E PREDICTED: hypothetical protein n=1 Tax=... 59 3e-07
UniRef100_UPI0000EDE64A PREDICTED: similar to smooth muscle myos... 59 3e-07
UniRef100_UPI0000EDE649 PREDICTED: similar to smooth muscle myos... 59 3e-07
UniRef100_UPI00006CD0F6 Protein kinase domain containing protein... 59 3e-07
UniRef100_UPI000058879A PREDICTED: similar to DNA-directed RNA p... 59 3e-07
UniRef100_UPI00006A19CB Kinesin-like protein KIF3C. n=1 Tax=Xeno... 59 3e-07
UniRef100_UPI0000D6352A smooth muscle myosin heavy chain 11 isof... 59 3e-07
UniRef100_Q8R384 Myh11 protein n=1 Tax=Mus musculus RepID=Q8R384... 59 3e-07
UniRef100_Q69ZX3 MKIAA0866 protein (Fragment) n=2 Tax=Mus muscul... 59 3e-07
UniRef100_Q2SR10 Putative uncharacterized protein n=1 Tax=Mycopl... 59 3e-07
UniRef100_C0MB36 Putative cell surface-anchored protein n=1 Tax=... 59 3e-07
UniRef100_A3IHQ1 Putative uncharacterized protein n=1 Tax=Cyanot... 59 3e-07
UniRef100_B9S0A4 ATSMC2, putative n=1 Tax=Ricinus communis RepID... 59 3e-07
UniRef100_Q9NJ21 Myosin heavy chain cardiac muscle specific isof... 59 3e-07
UniRef100_Q9NJ20 Myosin heavy chain cardiac muscle specific isof... 59 3e-07
UniRef100_Q8I5Y2 Conserved Plasmodium protein n=1 Tax=Plasmodium... 59 3e-07
UniRef100_Q54DR3 Actin binding protein n=1 Tax=Dictyostelium dis... 59 3e-07
UniRef100_Q17042 Myosin heavy chain n=2 Tax=Argopecten irradians... 59 3e-07
UniRef100_B6K8Q5 Putative uncharacterized protein n=2 Tax=Toxopl... 59 3e-07
UniRef100_A4HAX1 Putative uncharacterized protein (Fragment) n=1... 59 3e-07
UniRef100_A2FX23 Formin Homology 2 Domain containing protein n=1... 59 3e-07
UniRef100_A2EQA8 Putative uncharacterized protein n=1 Tax=Tricho... 59 3e-07
UniRef100_A2EPL2 Putative uncharacterized protein n=1 Tax=Tricho... 59 3e-07
UniRef100_A0EF40 Chromosome undetermined scaffold_92, whole geno... 59 3e-07
UniRef100_A0DWS3 Chromosome undetermined scaffold_67, whole geno... 59 3e-07
UniRef100_A0BUH1 Chromosome undetermined scaffold_129, whole gen... 59 3e-07
UniRef100_C7ZKZ9 Putative uncharacterized protein n=1 Tax=Nectri... 59 3e-07
UniRef100_C1GHL4 Fibronectin type III domain-containing protein ... 59 3e-07
UniRef100_B2W5E1 Predicted protein n=1 Tax=Pyrenophora tritici-r... 59 3e-07
UniRef100_Q3ISN7 DNA double-strand break repair rad50 ATPase n=1... 59 3e-07
UniRef100_O28714 Chromosome segregation protein (Smc1) n=1 Tax=A... 59 3e-07
UniRef100_C6A0L9 DNA double-strand break repair rad50 ATPase n=1... 59 3e-07
UniRef100_P24733 Myosin heavy chain, striated muscle n=2 Tax=Arg... 59 3e-07
UniRef100_O08638-2 Isoform 2 of Myosin-11 n=1 Tax=Mus musculus R... 59 3e-07
UniRef100_O08638 Myosin-11 n=1 Tax=Mus musculus RepID=MYH11_MOUSE 59 3e-07
UniRef100_UPI000194D504 PREDICTED: similar to Myosin-11 isoform ... 59 4e-07
UniRef100_UPI000194D4E6 PREDICTED: similar to Myosin-11 isoform ... 59 4e-07
UniRef100_UPI000194C9CE PREDICTED: thyroid hormone receptor inte... 59 4e-07
UniRef100_UPI000186EA28 Centromeric protein E, putative n=1 Tax=... 59 4e-07
UniRef100_UPI000186AF91 hypothetical protein BRAFLDRAFT_108728 n... 59 4e-07
UniRef100_UPI0001796BEC PREDICTED: similar to benzodiazapine rec... 59 4e-07
UniRef100_UPI000151ABFE hypothetical protein PGUG_00990 n=1 Tax=... 59 4e-07
UniRef100_UPI0000F2DBB5 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI0000F2B565 PREDICTED: similar to CENTRIOLIN n=1 Tax... 59 4e-07
UniRef100_UPI0000E4A173 PREDICTED: hypothetical protein, partial... 59 4e-07
UniRef100_UPI0000D94630 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A52C6 PREDICTED: similar to cingulin-like 1 n=... 59 4e-07
UniRef100_UPI00005A1097 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1096 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1095 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1094 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1093 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1092 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1091 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1090 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108F PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108E PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108D PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108C PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108B PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A108A PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1089 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1088 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1087 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1086 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1085 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1084 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1083 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1082 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1081 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A1080 PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A107F PREDICTED: similar to smooth muscle myos... 59 4e-07
UniRef100_UPI00005A107E PREDICTED: similar to smooth muscle myos... 59 4e-07