Tomato SBM DataBase
Annotation
Gene prediction and modeling were performed using an automatic gene assignment software that employs ab initio gene-finding and similarity search. The ab initio gene-finding software used in this pipeline included GeneMark.hmm and SNAP with the Arabidopsis thaliana and Lotus japonicus trained matrices. Similarity searches to detect potential protein-coding exons were performed using the BLASTX function of BLAST against the TrEMBL library. A total of 77,779 potential protein-coding genes have been assigned, of which 37,738 were identified as being related to transposable elements.
IDs for the predicted genes
The protein-encoding genes thus assigned have been denoted with an ID consisting of the contig name followed by a sequential number in an increment of 10 from one end to another. Note that these gene IDs should be regarded as temporal. The SBM sequence data are currently being integrated into the whole genome assembly data by the International Tomato Sequencing Consortium, and new standardized IDs will be provided in the future.
Codes for the predicted genes
-
 Codes for the gene prediction categories
Codes for the gene prediction categories
| /A |
genes that were predicted by two gene finding software, GeneMark.hmm and SNAP, and were similar to LGI or full length cDNA sequences from microTom.
|
| /B |
genes that were predicted by both of the software mentioned above but were not similar to LGI or full length cDNA sequences.
|
| /C |
genes that were predicted by either one of the software mentioned above and were similar to LGI or full length cDNA sequences.
|
| /D |
genes predicted by either one of the software mentioned above but were not similar to any of the known genes.
|
-
 Codes for the degree of gene coverage
Codes for the degree of gene coverage
| /nf |
sequences with significant similarities to the registered genes in the TrEMBL database that cover 70% or more of the coding regions.
|
| /p |
sequences with significant similarities to the registered genes in the TrEMBL database that cover less than 70% of the coding regions.
|
| no indication |
sequences with no significant similarities to the registered genes in the TrEMBL database.
|
-
 Other codes for the predicted genes
Other codes for the predicted genes
| /TE |
genes related to transposable elements.
|
| /partial |
genes lacking either the initiation or the termination codon as they are located at an contig end.
|
| /tRNA |
transfer RNA genes |
 Codes for the gene prediction categories
Codes for the gene prediction categories
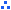 Codes for the gene prediction categories
Codes for the gene prediction categories
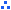 Codes for the degree of gene coverage
Codes for the degree of gene coverage
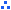 Other codes for the predicted genes
Other codes for the predicted genes