[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV410427 MWL072b07_r
(360 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
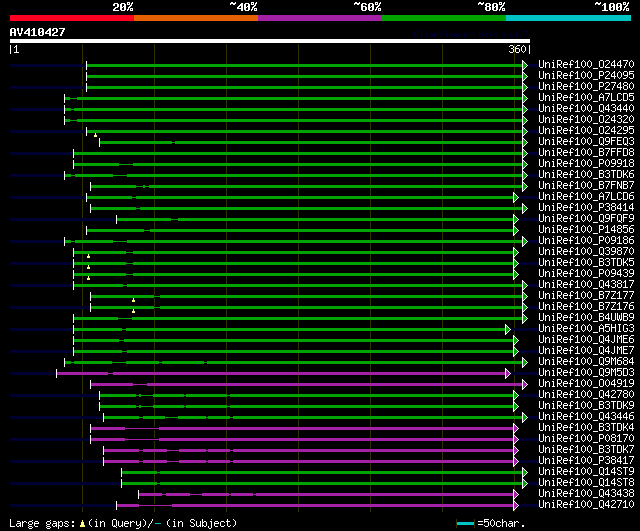
significant alignments:[graphical|details]
UniRef100_O24470 Lipoxygenase n=1 Tax=Pisum sativum RepID=O24470... 135 2e-30
UniRef100_P24095 Seed lipoxygenase n=1 Tax=Glycine max RepID=LOX... 133 5e-30
UniRef100_P27480 Lipoxygenase 1 n=1 Tax=Phaseolus vulgaris RepID... 130 6e-29
UniRef100_A7LCD5 Lipoxygenase n=1 Tax=Glycine max RepID=A7LCD5_S... 128 2e-28
UniRef100_Q43440 Lipoxygenase n=1 Tax=Glycine max RepID=Q43440_S... 125 1e-27
UniRef100_O24320 Lipoxygenase n=1 Tax=Phaseolus vulgaris RepID=O... 124 3e-27
UniRef100_O24295 Lipoxygenase n=1 Tax=Pisum sativum RepID=O24295... 119 1e-25
UniRef100_Q9FEQ3 Lipoxygenase n=1 Tax=Pisum sativum RepID=Q9FEQ3... 111 2e-23
UniRef100_B7FFD8 Putative uncharacterized protein (Fragment) n=1... 107 3e-22
UniRef100_P09918 Seed lipoxygenase-3 n=1 Tax=Pisum sativum RepID... 100 4e-20
UniRef100_B3TDK6 Lipoxygenase n=1 Tax=Glycine max RepID=B3TDK6_S... 100 5e-20
UniRef100_B7FNB7 Putative uncharacterized protein n=1 Tax=Medica... 100 7e-20
UniRef100_A7LCD6 Lipoxygenase n=1 Tax=Glycine max RepID=A7LCD6_S... 99 1e-19
UniRef100_P38414 Lipoxygenase n=1 Tax=Lens culinaris RepID=LOX1_... 99 2e-19
UniRef100_Q9FQF9 Lipoxygenase n=1 Tax=Phaseolus vulgaris RepID=Q... 97 4e-19
UniRef100_P14856 Seed lipoxygenase-2 n=1 Tax=Pisum sativum RepID... 97 4e-19
UniRef100_P09186 Seed lipoxygenase-3 n=1 Tax=Glycine max RepID=L... 96 1e-18
UniRef100_Q39870 Lipoxygenase n=1 Tax=Glycine max RepID=Q39870_S... 95 3e-18
UniRef100_B3TDK5 Lipoxygenase n=1 Tax=Glycine max RepID=B3TDK5_S... 95 3e-18
UniRef100_P09439 Seed lipoxygenase-2 n=1 Tax=Glycine max RepID=L... 95 3e-18
UniRef100_Q43817 Lipoxygenase n=1 Tax=Pisum sativum RepID=Q43817... 94 4e-18
UniRef100_B7Z177 Lipoxygenase n=1 Tax=Pisum sativum RepID=B7Z177... 92 2e-17
UniRef100_B7Z176 Lipoxygenase n=1 Tax=Pisum sativum RepID=B7Z176... 92 2e-17
UniRef100_B4UWB9 Lipoxygenase 1 (Fragment) n=1 Tax=Arachis hypog... 90 7e-17
UniRef100_A5HIG3 Lipoxygenase n=1 Tax=Caragana jubata RepID=A5HI... 86 2e-15
UniRef100_Q4JME6 Lipoxygenase n=1 Tax=Arachis hypogaea RepID=Q4J... 85 2e-15
UniRef100_Q4JME7 Lipoxygenase n=1 Tax=Arachis hypogaea RepID=Q4J... 83 8e-15
UniRef100_Q9M684 Lipoxygenase (Fragment) n=1 Tax=Phaseolus vulga... 82 2e-14
UniRef100_Q9M5D3 Lipoxygenase n=1 Tax=Arachis hypogaea RepID=Q9M... 79 1e-13
UniRef100_O04919 Lipoxygenase n=1 Tax=Vicia faba RepID=O04919_VICFA 79 2e-13
UniRef100_Q42780 Lipoxygenase n=1 Tax=Glycine max RepID=Q42780_S... 75 2e-12
UniRef100_B3TDK9 Lipoxygenase n=1 Tax=Glycine max RepID=B3TDK9_S... 75 2e-12
UniRef100_Q43446 Lipoxygenase n=1 Tax=Glycine max RepID=Q43446_S... 73 9e-12
UniRef100_B3TDK4 Lipoxygenase n=1 Tax=Glycine max RepID=B3TDK4_S... 73 1e-11
UniRef100_P08170 Seed lipoxygenase-1 n=1 Tax=Glycine max RepID=L... 73 1e-11
UniRef100_B3TDK7 Lipoxygenase n=1 Tax=Glycine max RepID=B3TDK7_S... 68 3e-10
UniRef100_P38417 Lipoxygenase-4 n=1 Tax=Glycine max RepID=LOX4_S... 68 3e-10
UniRef100_Q14ST9 Lipoxygenase n=1 Tax=Pisum sativum RepID=Q14ST9... 67 8e-10
UniRef100_Q14ST8 Lipoxygenase n=1 Tax=Pisum sativum RepID=Q14ST8... 67 8e-10
UniRef100_Q43438 Lipoxygenase n=1 Tax=Glycine max RepID=Q43438_S... 55 3e-06
UniRef100_Q42710 Lipoxygenase n=1 Tax=Cucumis sativus RepID=Q427... 54 5e-06
UniRef100_Q42704 Lipoxygenase n=1 Tax=Cucumis sativus RepID=Q427... 54 5e-06