[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
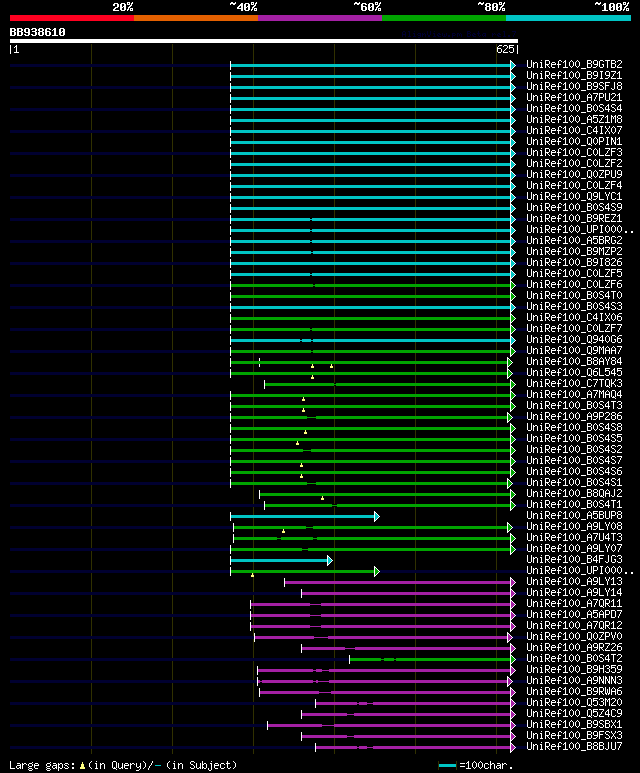
Query= BB938610 RCC11893
(625 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9GTB2 Predicted protein n=1 Tax=Populus trichocarpa R... 207 6e-52
UniRef100_B9I9Z1 Predicted protein n=1 Tax=Populus trichocarpa R... 206 1e-51
UniRef100_B9SFJ8 Gibberellin receptor GID1, putative n=1 Tax=Ric... 202 2e-50
UniRef100_A7PU21 Chromosome chr7 scaffold_31, whole genome shotg... 196 1e-48
UniRef100_B0S4S4 Putative GID1-like gibberellin receptor n=1 Tax... 195 2e-48
UniRef100_A5Z1M8 Gibberellic acid receptor-b n=1 Tax=Gossypium h... 195 2e-48
UniRef100_C4IX07 Putative gibberellin receptor n=1 Tax=Cucurbita... 192 2e-47
UniRef100_Q0PIN1 Gibberellic acid receptor n=1 Tax=Gossypium hir... 191 4e-47
UniRef100_C0LZF3 GID1-2 n=1 Tax=Gossypium hirsutum RepID=C0LZF3_... 191 4e-47
UniRef100_C0LZF2 GID1-1 n=1 Tax=Gossypium hirsutum RepID=C0LZF2_... 191 4e-47
UniRef100_Q0ZPU9 CXE carboxylesterase n=1 Tax=Actinidia delicios... 184 7e-45
UniRef100_C0LZF4 GID1-3 n=1 Tax=Gossypium hirsutum RepID=C0LZF4_... 182 3e-44
UniRef100_Q9LYC1 Probable gibberellin receptor GID1L2 n=1 Tax=Ar... 179 1e-43
UniRef100_B0S4S9 Putative GID1-like gibberellin receptor n=1 Tax... 176 1e-42
UniRef100_B9REZ1 Gibberellin receptor GID1, putative n=1 Tax=Ric... 171 5e-41
UniRef100_UPI0001984937 PREDICTED: hypothetical protein n=1 Tax=... 169 1e-40
UniRef100_A5BRG2 Chromosome chr14 scaffold_164, whole genome sho... 169 1e-40
UniRef100_B9MZP2 Predicted protein n=1 Tax=Populus trichocarpa R... 168 3e-40
UniRef100_B9I826 Predicted protein n=1 Tax=Populus trichocarpa R... 168 3e-40
UniRef100_C0LZF5 GID1-4 n=1 Tax=Gossypium hirsutum RepID=C0LZF5_... 167 5e-40
UniRef100_C0LZF6 GID1-5 n=1 Tax=Gossypium hirsutum RepID=C0LZF6_... 164 4e-39
UniRef100_B0S4T0 Putative GID1-like gibberellin receptor n=1 Tax... 163 9e-39
UniRef100_B0S4S3 Putative GID1-like gibberellin receptor n=1 Tax... 162 2e-38
UniRef100_C4IX06 Putative gibberellin receptor n=1 Tax=Cucurbita... 160 6e-38
UniRef100_C0LZF7 GID1-6 n=1 Tax=Gossypium hirsutum RepID=C0LZF7_... 160 6e-38
UniRef100_Q940G6 Probable gibberellin receptor GID1L3 n=1 Tax=Ar... 160 8e-38
UniRef100_Q9MAA7 Probable gibberellin receptor GID1L1 n=1 Tax=Ar... 150 6e-35
UniRef100_B8AY84 Putative uncharacterized protein n=1 Tax=Oryza ... 141 4e-32
UniRef100_Q6L545 Gibberellin receptor GID1 n=2 Tax=Oryza sativa ... 139 1e-31
UniRef100_C7TQK3 Putative GID1 protein (Fragment) n=1 Tax=Rosa l... 135 2e-30
UniRef100_A7MAQ4 GID1-like gibberellin receptor n=2 Tax=Hordeum ... 135 3e-30
UniRef100_B0S4T3 Putative GID1-like gibberellin receptor n=1 Tax... 133 1e-29
UniRef100_A9P286 Putative uncharacterized protein n=1 Tax=Picea ... 130 9e-29
UniRef100_B0S4S8 Putative GID1-like gibberellin receptor n=1 Tax... 129 3e-28
UniRef100_B0S4S5 Putative GID1-like gibberellin receptor n=1 Tax... 129 3e-28
UniRef100_B0S4S2 Putative GID1-like gibberellin receptor n=1 Tax... 127 8e-28
UniRef100_B0S4S7 Putative GID1-like gibberellin receptor n=1 Tax... 127 1e-27
UniRef100_B0S4S6 Putative GID1-like gibberellin receptor n=1 Tax... 127 1e-27
UniRef100_B0S4S1 Putative GID1-like gibberellin receptor n=1 Tax... 127 1e-27
UniRef100_B8QAJ2 Putative GA receptor GID1 (Fragment) n=1 Tax=Tr... 118 5e-25
UniRef100_B0S4T1 Putative GID1-like gibberellin receptor (Fragme... 110 1e-22
UniRef100_A5BUP8 Putative uncharacterized protein n=1 Tax=Vitis ... 108 5e-22
UniRef100_A9LY08 Putative gibberellin receptor n=1 Tax=Selaginel... 105 3e-21
UniRef100_A7U4T3 Putative gibberellin receptor n=1 Tax=Selaginel... 101 4e-20
UniRef100_A9LY07 Putative gibberellin receptor n=1 Tax=Selaginel... 99 3e-19
UniRef100_B4FJG3 Putative uncharacterized protein n=1 Tax=Zea ma... 72 3e-11
UniRef100_UPI00019858B4 PREDICTED: hypothetical protein n=1 Tax=... 70 1e-10
UniRef100_A9LY13 Gibberellin receptor GID1-like protein n=2 Tax=... 65 6e-09
UniRef100_A9LY14 Gibberellin receptor GID1-like protein n=1 Tax=... 64 1e-08
UniRef100_A7QR11 Chromosome undetermined scaffold_147, whole gen... 63 2e-08
UniRef100_A5APD7 Putative uncharacterized protein n=1 Tax=Vitis ... 63 2e-08
UniRef100_A7QR12 Chromosome undetermined scaffold_147, whole gen... 62 4e-08
UniRef100_Q0ZPV0 CXE carboxylesterase n=1 Tax=Actinidia delicios... 61 7e-08
UniRef100_A9RZ26 GLP2 GID1-like protein n=1 Tax=Physcomitrella p... 60 1e-07
UniRef100_B0S4T2 Putative GID1-like gibberellin receptor (Fragme... 59 4e-07
UniRef100_B9H359 Predicted protein n=1 Tax=Populus trichocarpa R... 58 7e-07
UniRef100_A9NNN3 Putative uncharacterized protein n=1 Tax=Picea ... 57 1e-06
UniRef100_B9RWA6 Gibberellin receptor GID1, putative n=1 Tax=Ric... 56 2e-06
UniRef100_Q53M20 Os11g0240600 protein n=2 Tax=Oryza sativa Japon... 56 3e-06
UniRef100_Q5Z4C9 Os06g0306600 protein n=1 Tax=Oryza sativa Japon... 55 4e-06
UniRef100_B9SBX1 Arylacetamide deacetylase, putative n=1 Tax=Ric... 55 4e-06
UniRef100_B9FSX3 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_B8BJU7 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_A2YC54 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06