[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
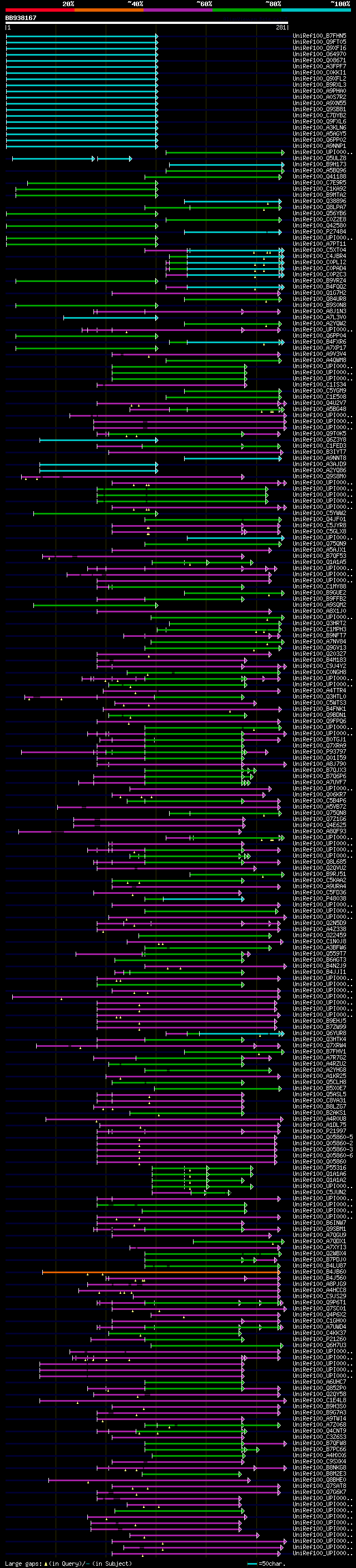
Query= BB938167 RCC11387
(281 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B7FHN5 Putative uncharacterized protein (Fragment) n=1... 108 2e-22
UniRef100_Q9FT05 Cationic peroxidase n=1 Tax=Cicer arietinum Rep... 103 6e-21
UniRef100_Q9XFI6 Putative uncharacterized protein n=1 Tax=Glycin... 103 8e-21
UniRef100_O64970 Putative uncharacterized protein n=1 Tax=Glycin... 103 8e-21
UniRef100_Q08671 Peroxidase n=1 Tax=Gossypium hirsutum RepID=Q08... 99 2e-19
UniRef100_A3FPF7 Cationic peroxidase n=1 Tax=Nelumbo nucifera Re... 98 2e-19
UniRef100_C0KKI1 Cationic peroxidase (Fragment) n=1 Tax=Tamarix ... 98 3e-19
UniRef100_Q9XFL2 Secretory peroxidase n=1 Tax=Nicotiana tabacum ... 97 6e-19
UniRef100_B9RXL3 Peroxidase 63, putative n=1 Tax=Ricinus communi... 95 3e-18
UniRef100_A9PHA0 Peroxidase n=1 Tax=Populus trichocarpa RepID=A9... 95 3e-18
UniRef100_A0S7R2 Class III peroxidase n=1 Tax=Oncidium Gower Ram... 95 3e-18
UniRef100_A9XN55 Peroxidase n=1 Tax=Thellungiella halophila RepI... 94 6e-18
UniRef100_Q9SB81 Peroxidase 42 n=1 Tax=Arabidopsis thaliana RepI... 94 6e-18
UniRef100_C7DYB2 Peroxidase n=1 Tax=Camellia oleifera RepID=C7DY... 93 8e-18
UniRef100_Q9FXL6 Secretory peroxidase n=1 Tax=Avicennia marina R... 92 1e-17
UniRef100_A3KLN6 Putative secretory peroxidase n=1 Tax=Catharant... 92 1e-17
UniRef100_A5AGY5 Chromosome chr10 scaffold_282, whole genome sho... 90 7e-17
UniRef100_Q6PP02 Peroxidase (Fragment) n=1 Tax=Mirabilis jalapa ... 88 3e-16
UniRef100_A9NNP1 Putative uncharacterized protein n=1 Tax=Picea ... 86 1e-15
UniRef100_UPI0001982DB4 PREDICTED: hypothetical protein n=1 Tax=... 72 2e-11
UniRef100_Q5ULZ8 Peroxidase (Fragment) n=1 Tax=Fragaria x ananas... 61 5e-11
UniRef100_B9H173 Predicted protein n=1 Tax=Populus trichocarpa R... 70 7e-11
UniRef100_A5BQ96 Putative uncharacterized protein n=1 Tax=Vitis ... 70 9e-11
UniRef100_Q41188 Glycine-rich protein n=1 Tax=Arabidopsis thalia... 67 8e-10
UniRef100_C7E9R5 Peroxidase 21 (Fragment) n=1 Tax=Brassica rapa ... 66 1e-09
UniRef100_C1KA92 Peroxidase n=1 Tax=Populus trichocarpa RepID=C1... 66 1e-09
UniRef100_B9MTA2 Predicted protein n=1 Tax=Populus trichocarpa R... 66 1e-09
UniRef100_Q38896 Glycine-rich protein 2b n=2 Tax=Arabidopsis tha... 66 1e-09
UniRef100_Q8LPA7 Cold shock protein-1 n=1 Tax=Triticum aestivum ... 66 1e-09
UniRef100_Q56YB6 Putative peroxidase ATP2a n=1 Tax=Arabidopsis t... 66 1e-09
UniRef100_C0Z2E8 AT4G38680 protein n=1 Tax=Arabidopsis thaliana ... 66 1e-09
UniRef100_Q42580 Peroxidase 21 n=2 Tax=Arabidopsis thaliana RepI... 66 1e-09
UniRef100_P27484 Glycine-rich protein 2 n=1 Tax=Nicotiana sylves... 65 2e-09
UniRef100_UPI0001983D26 PREDICTED: hypothetical protein n=1 Tax=... 65 2e-09
UniRef100_A7PT11 Chromosome chr8 scaffold_29, whole genome shotg... 65 2e-09
UniRef100_C5XT04 Putative uncharacterized protein Sb04g001720 n=... 64 4e-09
UniRef100_C4JBR4 Putative uncharacterized protein n=1 Tax=Zea ma... 64 4e-09
UniRef100_C0PLI2 Putative uncharacterized protein n=1 Tax=Zea ma... 64 4e-09
UniRef100_C0PAD4 Putative uncharacterized protein n=1 Tax=Zea ma... 64 4e-09
UniRef100_C0P2C3 Putative uncharacterized protein n=1 Tax=Zea ma... 64 4e-09
UniRef100_B9VRZ4 Peroxidase (Fragment) n=1 Tax=Cucumis sativus R... 64 4e-09
UniRef100_B4FQQ2 Putative uncharacterized protein n=1 Tax=Zea ma... 64 4e-09
UniRef100_Q1G7H2 Dishevelled-associated activator of morphogenes... 63 1e-08
UniRef100_Q84UR8 Os08g0129200 protein n=1 Tax=Oryza sativa Japon... 63 1e-08
UniRef100_B9S0N8 Peroxidase 21, putative n=1 Tax=Ricinus communi... 63 1e-08
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 63 1e-08
UniRef100_A7L3V0 Cationic peroxidase (Fragment) n=1 Tax=Phaseolu... 63 1e-08
UniRef100_A2YQW2 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_Q6PP04 Peroxidase (Fragment) n=1 Tax=Mirabilis jalapa ... 62 2e-08
UniRef100_B4FXR6 Putative uncharacterized protein n=1 Tax=Zea ma... 62 2e-08
UniRef100_A7XP17 Lignin peroxidase (Fragment) n=1 Tax=Garcinia m... 62 3e-08
UniRef100_A9V3V4 Predicted protein n=1 Tax=Monosiga brevicollis ... 62 3e-08
UniRef100_A4QWM8 Putative uncharacterized protein n=1 Tax=Magnap... 62 3e-08
UniRef100_UPI000194C8B0 PREDICTED: dishevelled associated activa... 61 3e-08
UniRef100_UPI000194C8AF PREDICTED: dishevelled associated activa... 61 3e-08
UniRef100_UPI000194C8AE PREDICTED: dishevelled associated activa... 61 3e-08
UniRef100_C1IS34 Minicollagen-1 n=1 Tax=Malo kingi RepID=C1IS34_... 61 3e-08
UniRef100_C5YGM9 Putative uncharacterized protein Sb06g029650 n=... 61 4e-08
UniRef100_C1E508 Predicted protein n=1 Tax=Micromonas sp. RCC299... 61 4e-08
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 60 6e-08
UniRef100_A5BG48 Putative uncharacterized protein n=1 Tax=Vitis ... 60 6e-08
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 60 7e-08
UniRef100_UPI00017600E2 PREDICTED: im:7158925 n=1 Tax=Danio reri... 60 7e-08
UniRef100_UPI0001A2C389 UPI0001A2C389 related cluster n=1 Tax=Da... 60 7e-08
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 60 7e-08
UniRef100_Q6Z3Y8 Class III peroxidase 116 n=3 Tax=Oryza sativa J... 60 7e-08
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 7e-08
UniRef100_B3IYT7 Chloroplast unusual positioning 1A n=1 Tax=Adia... 60 7e-08
UniRef100_A9NNT8 Putative uncharacterized protein n=1 Tax=Picea ... 60 7e-08
UniRef100_A3AJD9 Putative uncharacterized protein n=1 Tax=Oryza ... 60 7e-08
UniRef100_A2YQ86 Putative uncharacterized protein n=1 Tax=Oryza ... 60 7e-08
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 60 7e-08
UniRef100_UPI0000E800CF PREDICTED: similar to formin 2 n=1 Tax=G... 60 1e-07
UniRef100_UPI0001B7A6BD enabled homolog n=1 Tax=Rattus norvegicu... 60 1e-07
UniRef100_UPI0001B7A6BC enabled homolog n=1 Tax=Rattus norvegicu... 60 1e-07
UniRef100_UPI0001B7A6A7 enabled homolog n=1 Tax=Rattus norvegicu... 60 1e-07
UniRef100_UPI0000ECC8C2 Formin-2. n=1 Tax=Gallus gallus RepID=UP... 60 1e-07
UniRef100_C5YWW2 Putative uncharacterized protein Sb09g018150 n=... 60 1e-07
UniRef100_Q4JF01 Vasa homlogue n=1 Tax=Platynereis dumerilii Rep... 60 1e-07
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 60 1e-07
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 60 1e-07
UniRef100_UPI00019251A7 PREDICTED: similar to predicted protein ... 59 1e-07
UniRef100_Q75QN9 Cold shock domain protein 2 n=1 Tax=Triticum ae... 59 1e-07
UniRef100_A5AJX1 Putative uncharacterized protein n=1 Tax=Vitis ... 59 1e-07
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 59 1e-07
UniRef100_Q1A1A5 Forkhead box protein G1 n=1 Tax=Chlorocebus pyg... 40 1e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 59 2e-07
UniRef100_UPI0000D9E2E7 PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_UPI0000122FAC Hypothetical protein CBG05832 n=1 Tax=Ca... 59 2e-07
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 2e-07
UniRef100_B9GUE2 Predicted protein n=1 Tax=Populus trichocarpa R... 59 2e-07
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 59 2e-07
UniRef100_A9SQM2 Predicted protein n=1 Tax=Physcomitrella patens... 59 2e-07
UniRef100_A8X1J0 C. briggsae CBR-GRL-4 protein n=1 Tax=Caenorhab... 59 2e-07
UniRef100_UPI00019852F3 PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_Q3HRT2 Putative glycine-rich protein n=1 Tax=Picea gla... 59 2e-07
UniRef100_C1MPH3 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 2e-07
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 59 2e-07
UniRef100_A7NV84 Chromosome chr18 scaffold_1, whole genome shotg... 59 2e-07
UniRef100_Q9GV13 Vasa-related protein CnVAS1 n=1 Tax=Hydra magni... 59 2e-07
UniRef100_Q20327 Ground-like (Grd related) protein 4 n=1 Tax=Cae... 59 2e-07
UniRef100_B4M183 GJ24177 n=1 Tax=Drosophila virilis RepID=B4M183... 59 2e-07
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 59 2e-07
UniRef100_C0NGH8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 59 2e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI00003684DA PREDICTED: fas ligand n=1 Tax=Pan troglo... 58 3e-07
UniRef100_A4TTR4 Putative uncharacterized protein n=1 Tax=Magnet... 58 3e-07
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 58 3e-07
UniRef100_C5WTS3 Putative uncharacterized protein Sb01g043600 n=... 58 3e-07
UniRef100_B4FNK1 Putative uncharacterized protein n=1 Tax=Zea ma... 58 3e-07
UniRef100_Q9BDN1 Tumor necrosis factor ligand superfamily member... 58 3e-07
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 58 3e-07
UniRef100_UPI0001924586 PREDICTED: vasa-related protein CnVAS1 n... 58 4e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 58 4e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 58 4e-07
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 58 4e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 58 4e-07
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 58 4e-07
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 58 4e-07
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 58 4e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 58 4e-07
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 58 4e-07
UniRef100_UPI0001661EAE PREDICTED: similar to Putative acrosin-l... 57 5e-07
UniRef100_Q06KR7 Viral capsid associated protein n=1 Tax=Anticar... 57 5e-07
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 57 5e-07
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 57 5e-07
UniRef100_Q75QN8 Cold shock domain protein 3 n=1 Tax=Triticum ae... 57 5e-07
UniRef100_Q7Z1G6 RNA-binding protein 6 n=1 Tax=Trypanosoma cruzi... 57 5e-07
UniRef100_Q4E625 RNA-binding protein 6, putative n=1 Tax=Trypano... 57 5e-07
UniRef100_A8QF93 EBNA-2 nuclear protein, putative n=1 Tax=Brugia... 57 5e-07
UniRef100_UPI00019830CC PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 57 6e-07
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_UPI0001553749 PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 57 6e-07
UniRef100_Q2QVU2 Transposon protein, putative, CACTA, En/Spm sub... 57 6e-07
UniRef100_B9RJ51 Cold shock protein, putative n=1 Tax=Ricinus co... 57 6e-07
UniRef100_C5KAA2 Putative uncharacterized protein n=1 Tax=Perkin... 57 6e-07
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 57 6e-07
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 57 6e-07
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 57 6e-07
UniRef100_UPI0000E2530B PREDICTED: similar to mKIAA1205 protein ... 57 8e-07
UniRef100_UPI00016E37F6 UPI00016E37F6 related cluster n=1 Tax=Ta... 57 8e-07
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 57 8e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 57 8e-07
UniRef100_A4Z338 Putative Peptidase, Caspase-like domain and TPR... 57 8e-07
UniRef100_O22459 Hydroxyproline-rich glycoprotein gas29p n=1 Tax... 57 8e-07
UniRef100_C1N0J8 Predicted protein n=1 Tax=Micromonas pusilla CC... 57 8e-07
UniRef100_A3BFW6 Putative uncharacterized protein n=1 Tax=Oryza ... 57 8e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 57 8e-07
UniRef100_B6AGT3 Putative uncharacterized protein n=1 Tax=Crypto... 57 8e-07
UniRef100_B4N2J9 GK19249 n=1 Tax=Drosophila willistoni RepID=B4N... 57 8e-07
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 57 8e-07
UniRef100_UPI00017C2CD7 PREDICTED: similar to formin-like 2 n=1 ... 56 1e-06
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI0000547F81 PREDICTED: formin 1 n=1 Tax=Danio rerio ... 56 1e-06
UniRef100_UPI0001A2D80C UPI0001A2D80C related cluster n=1 Tax=Da... 56 1e-06
UniRef100_UPI0000566A27 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 56 1e-06
UniRef100_UPI0000566A26 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 56 1e-06
UniRef100_UPI000179F39E UPI000179F39E related cluster n=1 Tax=Bo... 56 1e-06
UniRef100_B9EHJ5 Fmn1 protein n=1 Tax=Mus musculus RepID=B9EHJ5_... 56 1e-06
UniRef100_B7ZW99 Fmn1 protein n=1 Tax=Mus musculus RepID=B7ZW99_... 56 1e-06
UniRef100_Q6YUR8 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 56 1e-06
UniRef100_Q7XRW4 OSJNBb0062H02.5 protein n=2 Tax=Oryza sativa Re... 56 1e-06
UniRef100_B7FHV1 Putative uncharacterized protein n=1 Tax=Medica... 56 1e-06
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 56 1e-06
UniRef100_A4RZU2 Predicted protein n=1 Tax=Ostreococcus lucimari... 56 1e-06
UniRef100_A2YHG8 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 56 1e-06
UniRef100_Q5CLH8 Protease n=1 Tax=Cryptosporidium hominis RepID=... 56 1e-06
UniRef100_B5X0E7 Vasa (Fragment) n=1 Tax=Capitella sp. I Grassle... 56 1e-06
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 56 1e-06
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 56 1e-06
UniRef100_B8LZG7 Formin 1,2/cappuccino, putative n=1 Tax=Talarom... 56 1e-06
UniRef100_B2AKS1 Translation initiation factor IF-2 n=1 Tax=Podo... 56 1e-06
UniRef100_A4R0U8 Predicted protein n=1 Tax=Magnaporthe grisea Re... 56 1e-06
UniRef100_A1DL75 Actin associated protein Wsp1, putative n=1 Tax... 56 1e-06
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 56 1e-06
UniRef100_Q05860-5 Isoform 5 of Formin-1 n=1 Tax=Mus musculus Re... 56 1e-06
UniRef100_Q05860-2 Isoform 2 of Formin-1 n=1 Tax=Mus musculus Re... 56 1e-06
UniRef100_Q05860-3 Isoform 3 of Formin-1 n=1 Tax=Mus musculus Re... 56 1e-06
UniRef100_Q05860-6 Isoform 6 of Formin-1 n=1 Tax=Mus musculus Re... 56 1e-06
UniRef100_Q05860 Formin-1 n=1 Tax=Mus musculus RepID=FMN1_MOUSE 56 1e-06
UniRef100_P55316 Forkhead box protein G1 n=1 Tax=Homo sapiens Re... 39 1e-06
UniRef100_Q1A1A6 Forkhead box protein G1 n=1 Tax=Cebus capucinus... 39 1e-06
UniRef100_Q1A1A2 Forkhead box protein G1 n=1 Tax=Equus burchelli... 39 1e-06
UniRef100_UPI0000D9BB94 PREDICTED: similar to Forkhead box prote... 39 1e-06
UniRef100_C5JUN2 Putative uncharacterized protein n=1 Tax=Ajello... 39 1e-06
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI0000EBDEB7 PREDICTED: similar to formin-like 1 n=1 ... 56 1e-06
UniRef100_UPI0000D9F56F PREDICTED: similar to Protein CXorf45 n=... 56 1e-06
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 56 1e-06
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 56 1e-06
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 56 1e-06
UniRef100_A7QGU9 Chromosome chr16 scaffold_94, whole genome shot... 56 1e-06
UniRef100_A7QDX1 Chromosome chr4 scaffold_83, whole genome shotg... 56 1e-06
UniRef100_A7XYI3 JXC1 n=1 Tax=Monodelphis domestica RepID=A7XYI3... 56 1e-06
UniRef100_Q2WBX4 Vasa protein isoform n=1 Tax=Platynereis dumeri... 56 1e-06
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 56 1e-06
UniRef100_B4LU87 GJ17267 n=1 Tax=Drosophila virilis RepID=B4LU87... 56 1e-06
UniRef100_B4JB60 GH10237 n=1 Tax=Drosophila grimshawi RepID=B4JB... 56 1e-06
UniRef100_B4J560 GH20878 n=1 Tax=Drosophila grimshawi RepID=B4J5... 56 1e-06
UniRef100_A8PJG9 Ground-like domain containing protein n=1 Tax=B... 56 1e-06
UniRef100_A4HCC8 Putative uncharacterized protein n=1 Tax=Leishm... 56 1e-06
UniRef100_C9JS29 Putative uncharacterized protein TNRC18 n=1 Tax... 56 1e-06
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 56 1e-06
UniRef100_Q7SC01 Putative uncharacterized protein n=1 Tax=Neuros... 56 1e-06
UniRef100_Q4P6X2 Putative uncharacterized protein n=1 Tax=Ustila... 56 1e-06
UniRef100_C1GH00 Cytokinesis protein sepA n=1 Tax=Paracoccidioid... 56 1e-06
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 56 1e-06
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 56 1e-06
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 56 1e-06
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 56 1e-06
UniRef100_UPI000194C22E PREDICTED: similar to formin 2 n=1 Tax=T... 55 2e-06
UniRef100_UPI0000DA3EC9 PREDICTED: similar to tumor endothelial ... 55 2e-06
UniRef100_UPI0001B7C13A Protein piccolo (Aczonin) (Multidomain p... 55 2e-06
UniRef100_UPI0001B7C139 Protein piccolo (Aczonin) (Multidomain p... 55 2e-06
UniRef100_UPI0001B7C138 Protein piccolo (Aczonin) (Multidomain p... 55 2e-06
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 55 2e-06
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 55 2e-06
UniRef100_Q2QY58 Transposon protein, putative, CACTA, En/Spm sub... 55 2e-06
UniRef100_C1E4L8 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 2e-06
UniRef100_B9H3S0 Predicted protein (Fragment) n=1 Tax=Populus tr... 55 2e-06
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 55 2e-06
UniRef100_A7Z068 LMOD2 protein n=1 Tax=Bos taurus RepID=A7Z068_B... 55 2e-06
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 55 2e-06
UniRef100_C3Z6S3 Putative uncharacterized protein n=1 Tax=Branch... 55 2e-06
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_A4HXX6 Hypothetical repeat protein n=1 Tax=Leishmania ... 55 2e-06
UniRef100_C9SXK4 Predicted protein n=1 Tax=Verticillium albo-atr... 55 2e-06
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 55 2e-06
UniRef100_B8M2E3 Adenylyl cyclase-associated protein n=1 Tax=Tal... 55 2e-06
UniRef100_Q8BHE0 Proline-rich protein 11 n=1 Tax=Mus musculus Re... 55 2e-06
UniRef100_Q7SAT8 Actin cytoskeleton-regulatory complex protein p... 55 2e-06
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 55 2e-06
UniRef100_UPI0001926BC7 PREDICTED: similar to mini-collagen isof... 55 2e-06
UniRef100_UPI0000F2E99A PREDICTED: similar to diaphanous homolog... 55 2e-06
UniRef100_UPI0000F2BFBF PREDICTED: similar to bromodomain PHD fi... 55 2e-06
UniRef100_UPI0000DA3E0C PREDICTED: similar to tumor endothelial ... 55 2e-06
UniRef100_UPI0000DA3A9F PREDICTED: similar to Formin-2 n=1 Tax=R... 55 2e-06
UniRef100_UPI0000603C6F PREDICTED: Ras association (RalGDS/AF-6)... 55 2e-06
UniRef100_UPI0001A2CA84 UPI0001A2CA84 related cluster n=1 Tax=Da... 55 2e-06
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 55 2e-06
UniRef100_UPI0001B79C20 additional sex combs like 3 (Drosophila)... 55 2e-06
UniRef100_UPI00015DF9DB Proline-rich protein 12. n=1 Tax=Homo sa... 55 2e-06
UniRef100_UPI0001596889 proline rich 12 n=1 Tax=Homo sapiens Rep... 55 2e-06
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 55 2e-06
UniRef100_UPI00005A148F PREDICTED: similar to Tumor necrosis fac... 55 2e-06
UniRef100_Q65553 UL36 n=3 Tax=Bovine herpesvirus 1 RepID=Q65553_... 55 2e-06
UniRef100_Q9WX60 Ccp protein n=1 Tax=Gluconacetobacter xylinus R... 55 2e-06
UniRef100_Q1NB62 OmpA/MotB n=1 Tax=Sphingomonas sp. SKA58 RepID=... 55 2e-06
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 55 2e-06
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 55 2e-06
UniRef100_Q5W6I0 Putative uncharacterized protein OSJNBb0115F21.... 55 2e-06
UniRef100_B9T6D3 Cellular nucleic acid binding protein, putative... 55 2e-06
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 55 2e-06
UniRef100_B9RYC5 Serine-threonine protein kinase, plant-type, pu... 55 2e-06
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 55 2e-06
UniRef100_B9FLY6 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_B8AWI6 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_B8A7W6 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_B7GE03 Predicted protein n=1 Tax=Phaeodactylum tricorn... 55 2e-06
UniRef100_A9U297 Predicted protein n=1 Tax=Physcomitrella patens... 55 2e-06
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 55 2e-06
UniRef100_B7PNV6 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B5DH56 GA25354 n=1 Tax=Drosophila pseudoobscura pseudo... 55 2e-06
UniRef100_A9URR1 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 2e-06
UniRef100_A8XTD1 Putative uncharacterized protein n=1 Tax=Caenor... 55 2e-06
UniRef100_A4H3P3 Putative uncharacterized protein n=1 Tax=Leishm... 55 2e-06
UniRef100_A6NFN6 Putative uncharacterized protein PRR12 n=1 Tax=... 55 2e-06
UniRef100_C0NIM8 Putative uncharacterized protein n=1 Tax=Ajello... 55 2e-06
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 55 2e-06
UniRef100_B2WMY2 Putative uncharacterized protein n=1 Tax=Pyreno... 55 2e-06
UniRef100_P63307 Tumor necrosis factor ligand superfamily member... 55 2e-06
UniRef100_Q9ULL5-2 Isoform 2 of Proline-rich protein 12 n=1 Tax=... 55 2e-06
UniRef100_Q9ULL5 Proline-rich protein 12 n=1 Tax=Homo sapiens Re... 55 2e-06
UniRef100_Q03173-5 Isoform 4 of Protein enabled homolog n=1 Tax=... 55 2e-06
UniRef100_Q03173-2 Isoform 1 of Protein enabled homolog n=1 Tax=... 55 2e-06
UniRef100_Q03173-4 Isoform 3 of Protein enabled homolog n=1 Tax=... 55 2e-06
UniRef100_Q03173 Protein enabled homolog n=1 Tax=Mus musculus Re... 55 2e-06
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 55 3e-06
UniRef100_UPI000155BA60 PREDICTED: similar to hCG2029577, partia... 55 3e-06
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 55 3e-06
UniRef100_UPI0000E4A382 PREDICTED: similar to DNA-dependent prot... 55 3e-06
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 55 3e-06
UniRef100_UPI0000E47F32 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0000E47360 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 55 3e-06
UniRef100_UPI00015DF6E5 enabled homolog (Drosophila) n=1 Tax=Mus... 55 3e-06
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 55 3e-06
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 55 3e-06
UniRef100_UPI0001951234 Ras-associated and pleckstrin homology d... 55 3e-06
UniRef100_UPI000057F721 UPI000057F721 related cluster n=1 Tax=Bo... 55 3e-06
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 55 3e-06
UniRef100_Q8PPF4 Putative uncharacterized protein n=1 Tax=Xantho... 55 3e-06
UniRef100_Q2QZM4 Transposon protein, putative, CACTA, En/Spm sub... 55 3e-06
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 55 3e-06
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 55 3e-06
UniRef100_C1E4Y1 Putative uncharacterized protein n=1 Tax=Microm... 55 3e-06
UniRef100_B8AKX7 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 55 3e-06
UniRef100_A5D7V0 PRR11 protein n=1 Tax=Bos taurus RepID=A5D7V0_B... 55 3e-06
UniRef100_Q4CV65 RNA-binding protein 6, putative n=1 Tax=Trypano... 55 3e-06
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_B4L024 GI11736 n=1 Tax=Drosophila mojavensis RepID=B4L... 55 3e-06
UniRef100_A8QDB8 Ground-like domain containing protein n=1 Tax=B... 55 3e-06
UniRef100_C9K0J5 Putative uncharacterized protein RAPH1 n=1 Tax=... 55 3e-06
UniRef100_B7Z847 cDNA FLJ52583 n=1 Tax=Homo sapiens RepID=B7Z847... 55 3e-06
UniRef100_B7Z804 cDNA FLJ61626 n=1 Tax=Homo sapiens RepID=B7Z804... 55 3e-06
UniRef100_B1AKD6 Asparagine-linked glycosylation 13 homolog (S. ... 55 3e-06
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 55 3e-06
UniRef100_B0CT66 RhoA GTPase effector DIA/Diaphanous n=1 Tax=Lac... 55 3e-06
UniRef100_O15417-2 Isoform 2 of Trinucleotide repeat-containing ... 55 3e-06
UniRef100_Q70E73 Ras-associated and pleckstrin homology domains-... 55 3e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 55 3e-06
UniRef100_UPI00019262BB PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI00017C3792 PREDICTED: similar to additional sex com... 54 4e-06
UniRef100_UPI00015B5B2E PREDICTED: similar to ENSANGP00000010144... 54 4e-06
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI000155CFFC PREDICTED: similar to diaphanous homolog... 54 4e-06
UniRef100_UPI0000F2B170 PREDICTED: similar to hCG2004723, n=1 Ta... 54 4e-06
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 54 4e-06
UniRef100_UPI00005EB969 PREDICTED: similar to SET binding protei... 54 4e-06
UniRef100_UPI00005A6092 PREDICTED: similar to multidomain presyn... 54 4e-06
UniRef100_UPI00005A3937 PREDICTED: similar to formin-like 2 isof... 54 4e-06
UniRef100_UPI00005A14C7 PREDICTED: similar to Enabled protein ho... 54 4e-06
UniRef100_UPI000022352F Hypothetical protein CBG15462 n=1 Tax=Ca... 54 4e-06
UniRef100_UPI00004D6F7D Formin-like protein 2 (Formin homology 2... 54 4e-06
UniRef100_UPI0000DC1294 UPI0000DC1294 related cluster n=1 Tax=Ra... 54 4e-06
UniRef100_UPI0000DA344E proline rich 11 n=1 Tax=Rattus norvegicu... 54 4e-06
UniRef100_UPI0001B798A6 UPI0001B798A6 related cluster n=1 Tax=Ho... 54 4e-06
UniRef100_UPI0001AE6B4C SET domain-containing protein 1B n=1 Tax... 54 4e-06
UniRef100_UPI00016E37F5 UPI00016E37F5 related cluster n=1 Tax=Ta... 54 4e-06
UniRef100_UPI000179D3E7 UPI000179D3E7 related cluster n=1 Tax=Bo... 54 4e-06
UniRef100_UPI0000ECD10B Zinc finger homeobox protein 4 (Zinc fin... 54 4e-06
UniRef100_Q7ZWT3 Sf1 protein n=1 Tax=Xenopus laevis RepID=Q7ZWT3... 54 4e-06
UniRef100_Q4RLQ7 Chromosome 10 SCAF15019, whole genome shotgun s... 54 4e-06
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 54 4e-06
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 54 4e-06
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 54 4e-06
UniRef100_Q33AC7 Transposon protein, putative, CACTA, En/Spm sub... 54 4e-06
UniRef100_Q2QWV3 Transposon protein, putative, CACTA, En/Spm sub... 54 4e-06
UniRef100_Q7XME8 OSJNBa0061G20.11 protein n=2 Tax=Oryza sativa R... 54 4e-06
UniRef100_Q00ZZ2 Membrane coat complex Retromer, subunit VPS5/SN... 54 4e-06
UniRef100_C5YJ27 Putative uncharacterized protein Sb07g006896 (F... 54 4e-06
UniRef100_C1MU93 Predicted protein n=1 Tax=Micromonas pusilla CC... 54 4e-06
UniRef100_B9N8Z7 Predicted protein (Fragment) n=2 Tax=Populus tr... 54 4e-06
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 54 4e-06
UniRef100_Q8IMS9 CG31439 n=1 Tax=Drosophila melanogaster RepID=Q... 54 4e-06
UniRef100_Q00484 Mini-collagen n=1 Tax=Hydra sp. RepID=Q00484_9CNID 54 4e-06
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 54 4e-06
UniRef100_B6AF23 Putative uncharacterized protein n=1 Tax=Crypto... 54 4e-06
UniRef100_B4ND74 GK24962 n=1 Tax=Drosophila willistoni RepID=B4N... 54 4e-06
UniRef100_B3S718 Putative uncharacterized protein n=1 Tax=Tricho... 54 4e-06
UniRef100_B2KTD4 Minicollagen 1 n=1 Tax=Clytia hemisphaerica Rep... 54 4e-06
UniRef100_A8XM89 Putative uncharacterized protein n=1 Tax=Caenor... 54 4e-06
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 54 4e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 54 4e-06
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 54 4e-06
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 54 4e-06
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 54 4e-06
UniRef100_Q9UPS6 Histone-lysine N-methyltransferase SETD1B n=1 T... 54 4e-06
UniRef100_Q96PY5-3 Isoform 2 of Formin-like protein 2 n=1 Tax=Ho... 54 4e-06
UniRef100_Q96PY5 Formin-like protein 2 n=1 Tax=Homo sapiens RepI... 54 4e-06
UniRef100_P10323 Acrosin heavy chain n=1 Tax=Homo sapiens RepID=... 54 4e-06
UniRef100_UPI0001926925 PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI00019259C5 PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI0001792240 PREDICTED: similar to formin 3 CG33556-P... 54 5e-06
UniRef100_UPI00015B42DB PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI0000E49A24 PREDICTED: similar to LOC495507 protein ... 54 5e-06
UniRef100_UPI0000E254CE PREDICTED: piccolo isoform 3 n=1 Tax=Pan... 54 5e-06
UniRef100_UPI0000E254CD PREDICTED: piccolo isoform 2 n=1 Tax=Pan... 54 5e-06
UniRef100_UPI0000E254CC PREDICTED: piccolo isoform 1 n=1 Tax=Pan... 54 5e-06
UniRef100_UPI0000E1F761 PREDICTED: formin-like 2 isoform 2 n=1 T... 54 5e-06
UniRef100_UPI0000E1F760 PREDICTED: formin-like 2 isoform 8 n=1 T... 54 5e-06
UniRef100_UPI0000DA452D PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI000024DCC5 zinc finger homeodomain 4 n=1 Tax=Mus mu... 54 5e-06
UniRef100_UPI00000216EF glyceraldehyde-3-phosphate dehydrogenase... 54 5e-06
UniRef100_UPI0001573469 piccolo isoform 1 n=1 Tax=Homo sapiens R... 54 5e-06
UniRef100_UPI000156FA8C piccolo isoform 2 n=1 Tax=Homo sapiens R... 54 5e-06
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 54 5e-06
UniRef100_UPI00016E98C1 UPI00016E98C1 related cluster n=1 Tax=Ta... 54 5e-06
UniRef100_UPI00016E98A2 UPI00016E98A2 related cluster n=1 Tax=Ta... 54 5e-06
UniRef100_UPI000184A096 Zinc finger homeobox protein 4 (Zinc fin... 54 5e-06
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 54 5e-06
UniRef100_Q2NNS2 1629capsid n=1 Tax=Hyphantria cunea nucleopolyh... 54 5e-06
UniRef100_Q0C0P5 OmpA family protein n=1 Tax=Hyphomonas neptuniu... 54 5e-06
UniRef100_Q0ANI5 OmpA/MotB domain protein n=1 Tax=Maricaulis mar... 54 5e-06
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 54 5e-06
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 54 5e-06
UniRef100_B4YB55 BimA n=1 Tax=Burkholderia pseudomallei RepID=B4... 54 5e-06
UniRef100_A3WEW6 Peptidoglycan-associated protein n=1 Tax=Erythr... 54 5e-06
UniRef100_Q7XHB6 Transposon protein, putative, CACTA, En/Spm sub... 54 5e-06
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 54 5e-06
UniRef100_Q337W7 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_C5YLU5 Putative uncharacterized protein Sb07g000890 n=... 54 5e-06
UniRef100_B9G5V4 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_B8BH08 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_A9TVK2 Predicted protein n=1 Tax=Physcomitrella patens... 54 5e-06
UniRef100_A2XXJ0 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_C0JIS9 Nematoblast-specific protein nb039a-sv9 (Fragme... 54 5e-06
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 54 5e-06
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 54 5e-06
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 54 5e-06
UniRef100_B4LHU8 GJ11408 n=1 Tax=Drosophila virilis RepID=B4LHU8... 54 5e-06
UniRef100_B4K7T5 GI10549 n=1 Tax=Drosophila mojavensis RepID=B4K... 54 5e-06
UniRef100_B4IT00 GE11236 n=1 Tax=Drosophila yakuba RepID=B4IT00_... 54 5e-06
UniRef100_B3N5G6 GG25361, isoform A n=1 Tax=Drosophila erecta Re... 54 5e-06
UniRef100_C9J504 Putative uncharacterized protein ENSP0000041021... 54 5e-06
UniRef100_A6NG74 Putative uncharacterized protein PCLO n=2 Tax=H... 54 5e-06
UniRef100_C5PHL5 Putative uncharacterized protein n=1 Tax=Coccid... 54 5e-06
UniRef100_C5FCB8 Epstein-Barr nuclear antigen 2 n=1 Tax=Microspo... 54 5e-06
UniRef100_B6Q8P3 Actin cortical patch assembly protein Pan1, put... 54 5e-06
UniRef100_B0DAJ0 Predicted protein n=1 Tax=Laccaria bicolor S238... 54 5e-06
UniRef100_Q9Z0G8-2 Isoform 2 of WAS/WASL-interacting protein fam... 54 5e-06
UniRef100_Q9Z0G8 WAS/WASL-interacting protein family member 3 n=... 54 5e-06
UniRef100_Q9QYX7-2 Isoform 2 of Protein piccolo n=1 Tax=Mus musc... 54 5e-06
UniRef100_Q9QYX7-3 Isoform 3 of Protein piccolo n=1 Tax=Mus musc... 54 5e-06
UniRef100_Q9QYX7-4 Isoform 4 of Protein piccolo n=1 Tax=Mus musc... 54 5e-06
UniRef100_Q9QYX7 Protein piccolo n=1 Tax=Mus musculus RepID=PCLO... 54 5e-06
UniRef100_Q9Y6V0-2 Isoform 2 of Protein piccolo n=1 Tax=Homo sap... 54 5e-06
UniRef100_Q9Y6V0 Protein piccolo n=1 Tax=Homo sapiens RepID=PCLO... 54 5e-06
UniRef100_UPI0001985FA6 PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0001926BAB PREDICTED: similar to mini-collagen n=1 ... 54 7e-06
UniRef100_UPI00017605EC PREDICTED: similar to formin 2 n=1 Tax=D... 54 7e-06
UniRef100_UPI000175F433 PREDICTED: similar to formin-like 1 n=1 ... 54 7e-06
UniRef100_UPI00015FF5B0 piccolo isoform 2 n=1 Tax=Mus musculus R... 54 7e-06
UniRef100_UPI00015FA08D piccolo isoform 1 n=1 Tax=Mus musculus R... 54 7e-06
UniRef100_UPI0001553827 PREDICTED: additional sex combs like 3 n... 54 7e-06
UniRef100_UPI0000F2C630 PREDICTED: similar to zinc finger homeod... 54 7e-06
UniRef100_UPI0000E249A7 PREDICTED: similar to PRR11 protein n=1 ... 54 7e-06
UniRef100_UPI0000E1F767 PREDICTED: formin-like 2 isoform 3 n=1 T... 54 7e-06
UniRef100_UPI0000E1F766 PREDICTED: formin-like 2 isoform 1 n=1 T... 54 7e-06
UniRef100_UPI0000E1F765 PREDICTED: formin-like 2 isoform 7 n=1 T... 54 7e-06
UniRef100_UPI0000E1F763 PREDICTED: formin-like 2 isoform 4 n=1 T... 54 7e-06
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 54 7e-06
UniRef100_UPI00005A1758 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI00005A1757 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI00005A1756 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI00005A1755 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI00005A1754 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI00005A1753 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_UPI0001A2D69A UPI0001A2D69A related cluster n=1 Tax=Da... 54 7e-06
UniRef100_UPI0001B7A397 jumonji domain containing 3 n=1 Tax=Ratt... 54 7e-06
UniRef100_UPI00016D3858 piccolo (presynaptic cytomatrix protein)... 54 7e-06
UniRef100_UPI00016D3827 piccolo (presynaptic cytomatrix protein)... 54 7e-06
UniRef100_UPI00015DF6C1 piccolo (presynaptic cytomatrix protein)... 54 7e-06
UniRef100_UPI00016E98C2 UPI00016E98C2 related cluster n=1 Tax=Ta... 54 7e-06
UniRef100_UPI00016E37F2 UPI00016E37F2 related cluster n=1 Tax=Ta... 54 7e-06
UniRef100_UPI00005A1D18 PREDICTED: hypothetical protein XP_84743... 54 7e-06
UniRef100_UPI00005A1752 PREDICTED: similar to dishevelled-associ... 54 7e-06
UniRef100_Q4THH6 Chromosome undetermined SCAF2934, whole genome ... 54 7e-06
UniRef100_Q4TES1 Chromosome undetermined SCAF5157, whole genome ... 54 7e-06
UniRef100_Q4RY48 Chromosome 3 SCAF14978, whole genome shotgun se... 54 7e-06
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 54 7e-06
UniRef100_Q73TB8 Putative uncharacterized protein n=1 Tax=Mycoba... 54 7e-06
UniRef100_B9LBU3 Putative uncharacterized protein n=1 Tax=Chloro... 54 7e-06
UniRef100_B8H5M9 Cell surface antigen Sca2 n=2 Tax=Caulobacter v... 54 7e-06
UniRef100_B0T2W0 OmpA/MotB domain protein n=1 Tax=Caulobacter sp... 54 7e-06
UniRef100_A9WHI6 Putative uncharacterized protein n=1 Tax=Chloro... 54 7e-06
UniRef100_A4FBY5 Putative uncharacterized protein n=1 Tax=Saccha... 54 7e-06
UniRef100_Q6F3B9 Os03g0786500 protein n=1 Tax=Oryza sativa Japon... 54 7e-06
UniRef100_Q53N23 Transposon protein, putative, CACTA, En/Spm sub... 54 7e-06
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 54 7e-06
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 54 7e-06
UniRef100_Q10CE1 GRF zinc finger family protein, expressed n=1 T... 54 7e-06
UniRef100_C5XW81 Putative uncharacterized protein Sb04g005115 n=... 54 7e-06
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 54 7e-06
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 54 7e-06
UniRef100_A2XD25 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_Q5CS67 Signal peptide containing large protein with pr... 54 7e-06
UniRef100_C5KZZ4 Putative uncharacterized protein n=1 Tax=Perkin... 54 7e-06
UniRef100_C5KJV5 Glycoprotein X, putative n=1 Tax=Perkinsus mari... 54 7e-06
UniRef100_B7PJF9 Menin, putative n=1 Tax=Ixodes scapularis RepID... 54 7e-06
UniRef100_B5DLQ4 GA23099 (Fragment) n=1 Tax=Drosophila pseudoobs... 54 7e-06
UniRef100_B4NPJ4 GK18088 n=1 Tax=Drosophila willistoni RepID=B4N... 54 7e-06
UniRef100_B4NLW8 GK10515 n=1 Tax=Drosophila willistoni RepID=B4N... 54 7e-06
UniRef100_B4KHD1 GI15288 n=1 Tax=Drosophila mojavensis RepID=B4K... 54 7e-06
UniRef100_B4HK28 GM25091 n=1 Tax=Drosophila sechellia RepID=B4HK... 54 7e-06
UniRef100_A7T6H4 Predicted protein (Fragment) n=1 Tax=Nematostel... 54 7e-06
UniRef100_A0BFK7 Chromosome undetermined scaffold_104, whole gen... 54 7e-06
UniRef100_Q0VHD7 CD95 ligand n=1 Tax=Homo sapiens RepID=Q0VHD7_H... 54 7e-06
UniRef100_A8K6X5 cDNA FLJ78680, highly similar to Homo sapiens d... 54 7e-06
UniRef100_C1GXM9 Cytokinesis protein sepA n=1 Tax=Paracoccidioid... 54 7e-06
UniRef100_Q8T8R1 CCHC-type zinc finger protein CG3800 n=1 Tax=Dr... 54 7e-06
UniRef100_P48023-2 Isoform 2 of Tumor necrosis factor ligand sup... 54 7e-06
UniRef100_P48023 Tumor necrosis factor ligand superfamily member... 54 7e-06
UniRef100_Q01851 POU domain, class 4, transcription factor 1 n=1... 54 7e-06
UniRef100_Q5NCY0 Lysine-specific demethylase 6B n=1 Tax=Mus musc... 54 7e-06
UniRef100_Q64467 Glyceraldehyde-3-phosphate dehydrogenase, testi... 54 7e-06
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 54 7e-06