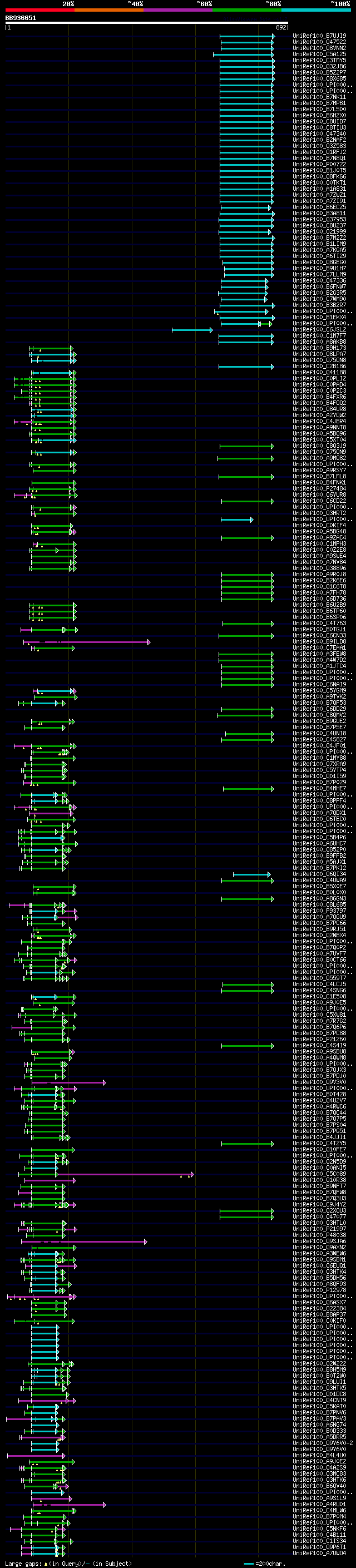
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB936651 RCC09623
(892 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B7UJI9 Beta-galactosidase n=1 Tax=Escherichia coli O12... 114 1e-23
UniRef100_Q47522 LacZ protein (Fragment) n=1 Tax=Escherichia col... 114 2e-23
UniRef100_Q8VNN2 Beta-galactosidase n=1 Tax=Escherichia coli Rep... 114 2e-23
UniRef100_C5A125 Tryptophan synthase alpha chain n=1 Tax=Escheri... 113 2e-23
UniRef100_C3TMY5 Beta-D-galactosidase n=1 Tax=Escherichia coli R... 112 4e-23
UniRef100_Q32JB6 Beta-galactosidase n=1 Tax=Shigella dysenteriae... 112 4e-23
UniRef100_B5Z2P7 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 112 4e-23
UniRef100_Q8X685 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 112 4e-23
UniRef100_UPI0001B5303F beta-D-galactosidase n=1 Tax=Shigella sp... 112 5e-23
UniRef100_UPI0001B525AD beta-D-galactosidase n=1 Tax=Escherichia... 112 5e-23
UniRef100_B7NK11 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 112 5e-23
UniRef100_B7MPB1 Beta-D-galactosidase n=1 Tax=Escherichia coli E... 112 5e-23
UniRef100_B7L500 Beta-D-galactosidase n=1 Tax=Escherichia coli 5... 112 5e-23
UniRef100_B6HZX0 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 112 5e-23
UniRef100_C8UID7 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 112 5e-23
UniRef100_C8TIU3 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 112 5e-23
UniRef100_Q47340 LacZ 5'-region (Fragment) n=2 Tax=Escherichia c... 112 5e-23
UniRef100_B2NAF2 Beta-galactosidase n=1 Tax=Escherichia coli 536... 112 5e-23
UniRef100_Q3Z583 Beta-galactosidase n=1 Tax=Shigella sonnei Ss04... 112 5e-23
UniRef100_Q1RFJ2 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 112 5e-23
UniRef100_B7N8Q1 Beta-galactosidase n=1 Tax=Escherichia coli UMN... 112 5e-23
UniRef100_P00722 Beta-galactosidase n=6 Tax=Escherichia coli Rep... 112 5e-23
UniRef100_B1J0T5 Beta-galactosidase n=1 Tax=Escherichia coli ATC... 112 5e-23
UniRef100_Q8FKG6 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 112 5e-23
UniRef100_Q0TKT1 Beta-galactosidase n=1 Tax=Escherichia coli 536... 112 5e-23
UniRef100_A1A831 Beta-galactosidase n=1 Tax=Escherichia coli APE... 112 5e-23
UniRef100_A7ZWZ1 Beta-galactosidase n=1 Tax=Escherichia coli HS ... 112 5e-23
UniRef100_A7ZI91 Beta-galactosidase n=1 Tax=Escherichia coli E24... 112 5e-23
UniRef100_B6ECZ5 LacZ alpha n=1 Tax=Cloning vector pMAK28 RepID=... 112 7e-23
UniRef100_B3A811 Beta-galactosidase n=1 Tax=Escherichia coli O15... 111 9e-23
UniRef100_Q37953 LacZ protein (Fragment) n=1 Tax=Phage M13mp18 R... 111 1e-22
UniRef100_C8U237 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 110 1e-22
UniRef100_O21999 LacZ-alpha n=1 Tax=Cloning vector pAL-Z RepID=O... 109 4e-22
UniRef100_B7M2Z2 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 108 6e-22
UniRef100_B1LIM9 Beta-galactosidase n=1 Tax=Escherichia coli SMS... 108 6e-22
UniRef100_A7KGA5 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 108 6e-22
UniRef100_A6TI29 Beta-galactosidase 2 n=1 Tax=Klebsiella pneumon... 108 6e-22
UniRef100_Q8GEG0 Putative uncharacterized protein n=1 Tax=Erwini... 108 1e-21
UniRef100_B9U1H7 Beta-D-galactosidase n=1 Tax=Vaccinia virus GLV... 107 2e-21
UniRef100_C7LLM9 Beta-galactosidase, chain D n=1 Tax=Mycoplasma ... 107 2e-21
UniRef100_Q47336 LacZ-alpha peptide n=1 Tax=Escherichia coli Rep... 105 6e-21
UniRef100_B6FNW7 Putative uncharacterized protein n=1 Tax=Clostr... 105 6e-21
UniRef100_B2G3R5 Putative puroindoline b protein n=1 Tax=Triticu... 105 8e-21
UniRef100_C7WM90 LacZ alpha protein (Fragment) n=1 Tax=Enterococ... 103 2e-20
UniRef100_B3B2R7 Beta-galactosidase n=1 Tax=Escherichia coli O15... 103 2e-20
UniRef100_UPI0000ECC20B UPI0000ECC20B related cluster n=1 Tax=Ga... 103 3e-20
UniRef100_B1EKX4 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 95 8e-18
UniRef100_UPI0000ECBAF8 UPI0000ECBAF8 related cluster n=1 Tax=Ga... 89 2e-16
UniRef100_C6JSL2 Putative uncharacterized protein Sb1292s002010 ... 90 3e-16
UniRef100_C1M7F7 Beta-D-galactosidase n=1 Tax=Citrobacter sp. 30... 90 4e-16
UniRef100_A8AKB8 Beta-galactosidase n=1 Tax=Citrobacter koseri A... 89 5e-16
UniRef100_B9H173 Predicted protein n=1 Tax=Populus trichocarpa R... 89 8e-16
UniRef100_Q8LPA7 Cold shock protein-1 n=1 Tax=Triticum aestivum ... 88 1e-15
UniRef100_Q75QN8 Cold shock domain protein 3 n=1 Tax=Triticum ae... 88 1e-15
UniRef100_C2B186 Putative uncharacterized protein n=1 Tax=Citrob... 86 4e-15
UniRef100_Q41188 Glycine-rich protein n=1 Tax=Arabidopsis thalia... 86 5e-15
UniRef100_C0PLI2 Putative uncharacterized protein n=1 Tax=Zea ma... 85 9e-15
UniRef100_C0PAD4 Putative uncharacterized protein n=1 Tax=Zea ma... 85 9e-15
UniRef100_C0P2C3 Putative uncharacterized protein n=1 Tax=Zea ma... 85 9e-15
UniRef100_B4FXR6 Putative uncharacterized protein n=1 Tax=Zea ma... 85 9e-15
UniRef100_B4FQQ2 Putative uncharacterized protein n=1 Tax=Zea ma... 85 9e-15
UniRef100_Q84UR8 Os08g0129200 protein n=1 Tax=Oryza sativa Japon... 84 2e-14
UniRef100_A2YQW2 Putative uncharacterized protein n=1 Tax=Oryza ... 84 2e-14
UniRef100_C4JBR4 Putative uncharacterized protein n=1 Tax=Zea ma... 83 3e-14
UniRef100_A9NNT8 Putative uncharacterized protein n=1 Tax=Picea ... 81 1e-13
UniRef100_A5BQ96 Putative uncharacterized protein n=1 Tax=Vitis ... 81 1e-13
UniRef100_C5XT04 Putative uncharacterized protein Sb04g001720 n=... 81 2e-13
UniRef100_C8Q3J9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 80 2e-13
UniRef100_Q75QN9 Cold shock domain protein 2 n=1 Tax=Triticum ae... 80 3e-13
UniRef100_A9MQ82 Beta-galactosidase n=1 Tax=Salmonella enterica ... 80 3e-13
UniRef100_UPI0001982DB4 PREDICTED: hypothetical protein n=1 Tax=... 79 5e-13
UniRef100_A9RSY7 Predicted protein n=1 Tax=Physcomitrella patens... 79 5e-13
UniRef100_B7LML8 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 77 2e-12
UniRef100_B4FNK1 Putative uncharacterized protein n=1 Tax=Zea ma... 77 2e-12
UniRef100_P27484 Glycine-rich protein 2 n=1 Tax=Nicotiana sylves... 77 2e-12
UniRef100_Q6YUR8 Putative uncharacterized protein n=1 Tax=Oryza ... 76 4e-12
UniRef100_C6CD22 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 76 5e-12
UniRef100_UPI00019852F3 PREDICTED: hypothetical protein n=1 Tax=... 75 7e-12
UniRef100_Q3HRT2 Putative glycine-rich protein n=1 Tax=Picea gla... 74 2e-11
UniRef100_UPI0000ECAFAB UPI0000ECAFAB related cluster n=1 Tax=Ga... 74 2e-11
UniRef100_C0KIF4 Vasa n=1 Tax=Strongylocentrotus purpuratus RepI... 74 2e-11
UniRef100_A5BG48 Putative uncharacterized protein n=1 Tax=Vitis ... 74 3e-11
UniRef100_A9ZAC4 Beta-galactosidase (Lactase) n=2 Tax=Yersinia p... 73 3e-11
UniRef100_C1MPH3 Predicted protein n=1 Tax=Micromonas pusilla CC... 73 3e-11
UniRef100_C0Z2E8 AT4G38680 protein n=1 Tax=Arabidopsis thaliana ... 73 3e-11
UniRef100_A9SWE4 Predicted protein n=1 Tax=Physcomitrella patens... 73 3e-11
UniRef100_A7NV84 Chromosome chr18 scaffold_1, whole genome shotg... 73 3e-11
UniRef100_Q38896 Glycine-rich protein 2b n=2 Tax=Arabidopsis tha... 73 3e-11
UniRef100_A9R0J8 Beta-galactosidase n=6 Tax=Yersinia pestis RepI... 73 3e-11
UniRef100_B2K6E6 Beta-galactosidase n=3 Tax=Yersinia pseudotuber... 73 3e-11
UniRef100_Q1C6T8 Beta-galactosidase n=8 Tax=Yersinia pestis RepI... 73 3e-11
UniRef100_A7FH78 Beta-galactosidase n=1 Tax=Yersinia pseudotuber... 73 3e-11
UniRef100_Q6D736 Beta-galactosidase n=1 Tax=Pectobacterium atros... 73 5e-11
UniRef100_B6U2B9 Glycine-rich protein 2b n=1 Tax=Zea mays RepID=... 72 6e-11
UniRef100_B6TP60 Glycine-rich protein 2b n=1 Tax=Zea mays RepID=... 72 6e-11
UniRef100_B6SP06 Glycine-rich protein 2b n=1 Tax=Zea mays RepID=... 72 6e-11
UniRef100_C4T763 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 8e-11
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 72 1e-10
UniRef100_C6CN33 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 1e-10
UniRef100_B9ILD8 Predicted protein (Fragment) n=1 Tax=Populus tr... 71 1e-10
UniRef100_C7EAA1 Vasa-like protein n=1 Tax=Haliotis asinina RepI... 71 1e-10
UniRef100_A3FEW8 Beta-galactosidase n=1 Tax=Pantoea agglomerans ... 71 1e-10
UniRef100_A4W7D2 Beta-galactosidase n=1 Tax=Enterobacter sp. 638... 71 1e-10
UniRef100_A1JTC4 Beta-galactosidase n=1 Tax=Yersinia enterocolit... 71 2e-10
UniRef100_UPI0001A43550 beta-D-galactosidase n=1 Tax=Pectobacter... 70 2e-10
UniRef100_UPI0001826C8D hypothetical protein ENTCAN_01162 n=1 Ta... 70 2e-10
UniRef100_C6NAI9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 2e-10
UniRef100_C5YGM9 Putative uncharacterized protein Sb06g029650 n=... 70 2e-10
UniRef100_A9TVK2 Predicted protein n=1 Tax=Physcomitrella patens... 70 2e-10
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 70 3e-10
UniRef100_C6DD29 Beta-galactosidase n=1 Tax=Pectobacterium carot... 70 3e-10
UniRef100_C8QMV2 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 3e-10
UniRef100_B9GUE2 Predicted protein n=1 Tax=Populus trichocarpa R... 70 3e-10
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 70 4e-10
UniRef100_C4UNI8 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 4e-10
UniRef100_C4S827 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 4e-10
UniRef100_Q4JF01 Vasa homlogue n=1 Tax=Platynereis dumerilii Rep... 70 4e-10
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 69 5e-10
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 69 5e-10
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 69 6e-10
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 69 6e-10
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 69 6e-10
UniRef100_B7P029 Vasa n=1 Tax=Chlamys farreri RepID=B7P029_9BIVA 69 7e-10
UniRef100_B4MHE7 GJ14580 n=1 Tax=Drosophila virilis RepID=B4MHE7... 69 7e-10
UniRef100_UPI00005EB969 PREDICTED: similar to SET binding protei... 69 8e-10
UniRef100_Q8PPF4 Putative uncharacterized protein n=1 Tax=Xantho... 69 8e-10
UniRef100_UPI00019830CC PREDICTED: hypothetical protein n=1 Tax=... 69 9e-10
UniRef100_A7QDX1 Chromosome chr4 scaffold_83, whole genome shotg... 69 9e-10
UniRef100_Q6TEC0 Vasa-like protein n=1 Tax=Crassostrea gigas Rep... 69 9e-10
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 68 1e-09
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 68 1e-09
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 68 1e-09
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 68 1e-09
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 68 1e-09
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 68 1e-09
UniRef100_A5AJX1 Putative uncharacterized protein n=1 Tax=Vitis ... 68 1e-09
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 68 1e-09
UniRef100_Q6QI34 LRRGT00174 n=1 Tax=Rattus norvegicus RepID=Q6QI... 68 1e-09
UniRef100_C4UWA9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 68 1e-09
UniRef100_B5X0E7 Vasa (Fragment) n=1 Tax=Capitella sp. I Grassle... 68 1e-09
UniRef100_B0L0X0 VASA n=1 Tax=Fenneropenaeus chinensis RepID=B0L... 68 1e-09
UniRef100_A8GGN3 Beta-galactosidase n=1 Tax=Serratia proteamacul... 68 1e-09
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 68 1e-09
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 68 1e-09
UniRef100_A7QGU9 Chromosome chr16 scaffold_94, whole genome shot... 68 1e-09
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 68 1e-09
UniRef100_B9RJ51 Cold shock protein, putative n=1 Tax=Ricinus co... 68 1e-09
UniRef100_Q2WBX4 Vasa protein isoform n=1 Tax=Platynereis dumeri... 68 1e-09
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 67 2e-09
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 67 2e-09
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 67 2e-09
UniRef100_B0CT66 RhoA GTPase effector DIA/Diaphanous n=1 Tax=Lac... 67 2e-09
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 67 2e-09
UniRef100_UPI0001553749 PREDICTED: hypothetical protein n=1 Tax=... 67 2e-09
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 67 2e-09
UniRef100_C4LCJ5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 2e-09
UniRef100_C4SNG6 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 2e-09
UniRef100_C1E508 Predicted protein n=1 Tax=Micromonas sp. RCC299... 67 2e-09
UniRef100_A9J0E5 DEAD box helicase n=1 Tax=Macrostomum lignano R... 67 2e-09
UniRef100_UPI0000F2BFBF PREDICTED: similar to bromodomain PHD fi... 67 3e-09
UniRef100_C5XW81 Putative uncharacterized protein Sb04g005115 n=... 67 3e-09
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 67 3e-09
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 67 3e-09
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 67 3e-09
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 67 3e-09
UniRef100_C4S4I9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 67 3e-09
UniRef100_A9SBU8 Predicted protein n=1 Tax=Physcomitrella patens... 67 3e-09
UniRef100_A4QWM8 Putative uncharacterized protein n=1 Tax=Magnap... 67 3e-09
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 66 4e-09
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 66 4e-09
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 66 4e-09
UniRef100_Q9V3V0 Xl6 n=1 Tax=Drosophila melanogaster RepID=Q9V3V... 66 4e-09
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 66 5e-09
UniRef100_B0T428 Glycoside hydrolase family 16 n=1 Tax=Caulobact... 66 5e-09
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 66 5e-09
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 66 5e-09
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 66 5e-09
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 66 5e-09
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 66 5e-09
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 66 5e-09
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 66 5e-09
UniRef100_C4TZY5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 66 6e-09
UniRef100_Q10FE7 Os03g0670700 protein n=1 Tax=Oryza sativa Japon... 66 6e-09
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 65 7e-09
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 65 7e-09
UniRef100_Q0ANI5 OmpA/MotB domain protein n=1 Tax=Maricaulis mar... 65 7e-09
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 65 7e-09
UniRef100_Q10R38 Putative uncharacterized protein n=1 Tax=Oryza ... 65 7e-09
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 65 7e-09
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 65 7e-09
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 65 7e-09
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 65 7e-09
UniRef100_Q2XQU3 Beta-galactosidase 2 n=1 Tax=Enterobacter cloac... 65 7e-09
UniRef100_Q47077 Beta-galactosidase n=1 Tax=Enterobacter cloacae... 65 7e-09
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 65 9e-09
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 65 9e-09
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 65 9e-09
UniRef100_Q9SJA6 Putative RSZp22 splicing factor n=1 Tax=Arabido... 65 9e-09
UniRef100_Q9AXN2 RNA-binding protein n=1 Tax=Triticum aestivum R... 65 9e-09
UniRef100_A3WEW6 Peptidoglycan-associated protein n=1 Tax=Erythr... 65 1e-08
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 65 1e-08
UniRef100_Q6EUQ1 Os02g0176100 protein n=1 Tax=Oryza sativa Japon... 65 1e-08
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 65 1e-08
UniRef100_B5DH56 GA25354 n=1 Tax=Drosophila pseudoobscura pseudo... 65 1e-08
UniRef100_A8QF93 EBNA-2 nuclear protein, putative n=1 Tax=Brugia... 65 1e-08
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 65 1e-08
UniRef100_UPI0000E47A48 PREDICTED: similar to HEXBP DNA binding ... 65 1e-08
UniRef100_Q6ASX7 Glycine-rich RNA binding protein n=1 Tax=Oryza ... 65 1e-08
UniRef100_O22384 Glycine-rich protein n=1 Tax=Oryza sativa RepID... 65 1e-08
UniRef100_B8AP37 Putative uncharacterized protein n=1 Tax=Oryza ... 65 1e-08
UniRef100_C0KIF0 Vasa n=1 Tax=Asterias forbesi RepID=C0KIF0_ASTFO 65 1e-08
UniRef100_UPI0000E254CE PREDICTED: piccolo isoform 3 n=1 Tax=Pan... 64 2e-08
UniRef100_UPI0000E254CD PREDICTED: piccolo isoform 2 n=1 Tax=Pan... 64 2e-08
UniRef100_UPI0000E254CC PREDICTED: piccolo isoform 1 n=1 Tax=Pan... 64 2e-08
UniRef100_UPI00005A6092 PREDICTED: similar to multidomain presyn... 64 2e-08
UniRef100_UPI0001573469 piccolo isoform 1 n=1 Tax=Homo sapiens R... 64 2e-08
UniRef100_UPI000156FA8C piccolo isoform 2 n=1 Tax=Homo sapiens R... 64 2e-08
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 64 2e-08
UniRef100_B8H5M9 Cell surface antigen Sca2 n=2 Tax=Caulobacter v... 64 2e-08
UniRef100_B0T2W0 OmpA/MotB domain protein n=1 Tax=Caulobacter sp... 64 2e-08
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 64 2e-08
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 64 2e-08
UniRef100_Q01DC8 Plg protein (ISS) n=1 Tax=Ostreococcus tauri Re... 64 2e-08
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 64 2e-08
UniRef100_C5KAT0 Putative uncharacterized protein n=1 Tax=Perkin... 64 2e-08
UniRef100_B7PNV6 Putative uncharacterized protein (Fragment) n=1... 64 2e-08
UniRef100_B7PAV3 Putative uncharacterized protein (Fragment) n=1... 64 2e-08
UniRef100_A6NG74 Putative uncharacterized protein PCLO n=2 Tax=H... 64 2e-08
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 64 2e-08
UniRef100_A5DRR5 Putative uncharacterized protein n=1 Tax=Lodder... 64 2e-08
UniRef100_Q9Y6V0-2 Isoform 2 of Protein piccolo n=1 Tax=Homo sap... 64 2e-08
UniRef100_Q9Y6V0 Protein piccolo n=1 Tax=Homo sapiens RepID=PCLO... 64 2e-08
UniRef100_B4L4U0 GI15829 n=1 Tax=Drosophila mojavensis RepID=B4L... 64 2e-08
UniRef100_A9J0E2 DEAD box helicase n=1 Tax=Macrostomum lignano R... 64 2e-08
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 64 2e-08
UniRef100_Q3MC83 Putative uncharacterized protein n=1 Tax=Anabae... 64 2e-08
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 64 2e-08
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 64 2e-08
UniRef100_UPI0000E468D6 PREDICTED: similar to laminin alpha chai... 64 2e-08
UniRef100_A9S1L9 Predicted protein n=1 Tax=Physcomitrella patens... 64 2e-08
UniRef100_A4RU01 Predicted protein n=1 Tax=Ostreococcus lucimari... 64 2e-08
UniRef100_C4MLW6 Vasa protein n=1 Tax=Parhyale hawaiensis RepID=... 64 2e-08
UniRef100_B7P0M4 Cement protein, putative (Fragment) n=1 Tax=Ixo... 64 2e-08
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 64 3e-08
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 64 3e-08
UniRef100_C4B111 Intracellular motility protein A n=4 Tax=Burkho... 64 3e-08
UniRef100_C1IS34 Minicollagen-1 n=1 Tax=Malo kingi RepID=C1IS34_... 64 3e-08
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 64 3e-08
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 64 3e-08
UniRef100_Q062H8 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 64 3e-08
UniRef100_C4QPC6 Cellular nucleic acid binding protein, putative... 64 3e-08
UniRef100_C4QPC5 Cellular nucleic acid binding protein, putative... 64 3e-08
UniRef100_UPI0001985FA6 PREDICTED: hypothetical protein n=1 Tax=... 63 4e-08
UniRef100_UPI0000DB6FAC PREDICTED: similar to Protein cappuccino... 63 4e-08
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 63 4e-08
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 63 4e-08
UniRef100_A5P9Z0 Peptidoglycan-associated protein n=1 Tax=Erythr... 63 4e-08
UniRef100_A7Z068 LMOD2 protein n=1 Tax=Bos taurus RepID=A7Z068_B... 63 4e-08
UniRef100_B3RJQ3 Putative uncharacterized protein n=1 Tax=Tricho... 63 4e-08
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 63 4e-08
UniRef100_UPI00005A3A5F PREDICTED: similar to zinc finger protei... 63 4e-08
UniRef100_A2C5L0 RNA-binding region RNP-1 (RNA recognition motif... 63 4e-08
UniRef100_C9XWJ2 Beta-galactosidase n=1 Tax=Cronobacter turicens... 63 4e-08
UniRef100_C4J159 Putative uncharacterized protein n=1 Tax=Zea ma... 63 4e-08
UniRef100_B9T6D3 Cellular nucleic acid binding protein, putative... 63 4e-08
UniRef100_B2VHN8 Beta-galactosidase n=1 Tax=Erwinia tasmaniensis... 63 4e-08
UniRef100_UPI0001661EAE PREDICTED: similar to Putative acrosin-l... 63 5e-08
UniRef100_B1WBQ8 Glyceraldehyde 3-phosphate dehydrogenase n=1 Ta... 63 5e-08
UniRef100_Q1RH03 Cell surface antigen Sca2 n=2 Tax=Rickettsia be... 63 5e-08
UniRef100_Q3L8Q3 Surface antigen (Fragment) n=1 Tax=Rickettsia b... 63 5e-08
UniRef100_B5FWY8 OmpA family protein n=1 Tax=Riemerella anatipes... 63 5e-08
UniRef100_Q3ECQ3 Uncharacterized protein At1g54215.1 n=1 Tax=Ara... 63 5e-08
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 63 5e-08
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 63 5e-08
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 63 5e-08
UniRef100_Q17G68 Formin 1,2/cappuccino n=1 Tax=Aedes aegypti Rep... 63 5e-08
UniRef100_B4QT65 GD21099 n=1 Tax=Drosophila simulans RepID=B4QT6... 63 5e-08
UniRef100_A8NHF1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 63 5e-08
UniRef100_Q9ESV6 Glyceraldehyde-3-phosphate dehydrogenase, testi... 63 5e-08
UniRef100_Q2QKC3 Pre-mRNA processing factor n=1 Tax=Triticum aes... 63 5e-08
UniRef100_C6TFM2 Putative uncharacterized protein n=1 Tax=Glycin... 63 5e-08
UniRef100_B9N668 Predicted protein n=1 Tax=Populus trichocarpa R... 63 5e-08
UniRef100_A8J4I2 SR protein factor n=1 Tax=Chlamydomonas reinhar... 63 5e-08
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 62 6e-08
UniRef100_Q2N609 Peptidoglycan-associated protein n=1 Tax=Erythr... 62 6e-08
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 62 6e-08
UniRef100_B7PJF9 Menin, putative n=1 Tax=Ixodes scapularis RepID... 62 6e-08
UniRef100_B7Z847 cDNA FLJ52583 n=1 Tax=Homo sapiens RepID=B7Z847... 62 6e-08
UniRef100_B7Z804 cDNA FLJ61626 n=1 Tax=Homo sapiens RepID=B7Z804... 62 6e-08
UniRef100_B1AKD6 Asparagine-linked glycosylation 13 homolog (S. ... 62 6e-08
UniRef100_UPI0000E4A204 PREDICTED: similar to zinc finger protei... 62 6e-08
UniRef100_UPI0000E49DCE PREDICTED: hypothetical protein n=1 Tax=... 62 6e-08
UniRef100_C1ZIK1 RRM domain-containing RNA-binding protein n=1 T... 62 6e-08
UniRef100_Q7XI13 Os07g0187300 protein n=1 Tax=Oryza sativa Japon... 62 6e-08
UniRef100_O81126 9G8-like SR protein n=1 Tax=Arabidopsis thalian... 62 6e-08
UniRef100_B8B7X7 Putative uncharacterized protein n=1 Tax=Oryza ... 62 6e-08
UniRef100_A8JAG8 Argonaute-like protein n=1 Tax=Chlamydomonas re... 62 6e-08
UniRef100_A7T8H9 Predicted protein (Fragment) n=1 Tax=Nematostel... 62 6e-08
UniRef100_A7MN76 Beta-galactosidase n=1 Tax=Cronobacter sakazaki... 62 6e-08
UniRef100_UPI000194BDE1 PREDICTED: zinc finger homeodomain 4 n=1... 62 8e-08
UniRef100_UPI0000E20C65 PREDICTED: similar to Family with sequen... 62 8e-08
UniRef100_UPI000069FBE4 Formin-like protein 2 (Formin homology 2... 62 8e-08
UniRef100_UPI0001B7C13A Protein piccolo (Aczonin) (Multidomain p... 62 8e-08
UniRef100_UPI0001B7C139 Protein piccolo (Aczonin) (Multidomain p... 62 8e-08
UniRef100_UPI0001B7C138 Protein piccolo (Aczonin) (Multidomain p... 62 8e-08
UniRef100_A3WBR8 Peptidoglycan-associated protein n=1 Tax=Erythr... 62 8e-08
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 62 8e-08
UniRef100_C1E4Y1 Putative uncharacterized protein n=1 Tax=Microm... 62 8e-08
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 62 8e-08
UniRef100_B4G6U6 GL19111 n=1 Tax=Drosophila persimilis RepID=B4G... 62 8e-08
UniRef100_UPI0000DC1358 UPI0000DC1358 related cluster n=1 Tax=Ra... 62 8e-08
UniRef100_Q31DH1 RNA-binding region RNP-1 n=1 Tax=Prochlorococcu... 62 8e-08
UniRef100_C5CSB2 RNP-1 like RNA-binding protein n=1 Tax=Variovor... 62 8e-08
UniRef100_C8TA13 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 62 8e-08
UniRef100_C4X846 Beta-D-galactosidase n=1 Tax=Klebsiella pneumon... 62 8e-08
UniRef100_B8B879 Putative uncharacterized protein n=1 Tax=Oryza ... 62 8e-08
UniRef100_A7VM16 Vasa-related protein n=1 Tax=Enchytraeus japone... 62 8e-08
UniRef100_A7RF28 Predicted protein n=1 Tax=Nematostella vectensi... 62 8e-08
UniRef100_Q6Z4K6 DEAD-box ATP-dependent RNA helicase 52B n=2 Tax... 62 8e-08
UniRef100_A6T8X0 Beta-galactosidase 1 n=1 Tax=Klebsiella pneumon... 62 8e-08
UniRef100_UPI0001553827 PREDICTED: additional sex combs like 3 n... 62 1e-07
UniRef100_UPI0000E23E7D PREDICTED: WAS protein homology region 2... 62 1e-07
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 62 1e-07
UniRef100_UPI00005A148F PREDICTED: similar to Tumor necrosis fac... 62 1e-07
UniRef100_Q4TB69 Chromosome 11 SCAF7190, whole genome shotgun se... 62 1e-07
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 62 1e-07
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 62 1e-07
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 62 1e-07
UniRef100_B9RYC5 Serine-threonine protein kinase, plant-type, pu... 62 1e-07
UniRef100_B4NYZ4 GE18300 n=1 Tax=Drosophila yakuba RepID=B4NYZ4_... 62 1e-07
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 62 1e-07
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 62 1e-07
UniRef100_Q8C4A5 Putative Polycomb group protein ASXL3 n=2 Tax=M... 62 1e-07
UniRef100_UPI0000DBF4CE UPI0000DBF4CE related cluster n=1 Tax=Ra... 62 1e-07
UniRef100_C5C660 Single-stranded DNA-binding protein n=1 Tax=Beu... 62 1e-07
UniRef100_B9QIA3 Putative uncharacterized protein n=1 Tax=Toxopl... 62 1e-07
UniRef100_B6KPI0 Putative uncharacterized protein n=1 Tax=Toxopl... 62 1e-07
UniRef100_UPI00015FF5B0 piccolo isoform 2 n=1 Tax=Mus musculus R... 61 1e-07
UniRef100_UPI00015FA08D piccolo isoform 1 n=1 Tax=Mus musculus R... 61 1e-07
UniRef100_UPI00016D3858 piccolo (presynaptic cytomatrix protein)... 61 1e-07
UniRef100_UPI00016D3827 piccolo (presynaptic cytomatrix protein)... 61 1e-07
UniRef100_UPI00015DF6C1 piccolo (presynaptic cytomatrix protein)... 61 1e-07
UniRef100_UPI00016E72DF UPI00016E72DF related cluster n=1 Tax=Ta... 61 1e-07
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 61 1e-07
UniRef100_Q0C0P5 OmpA family protein n=1 Tax=Hyphomonas neptuniu... 61 1e-07
UniRef100_A4TEZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 61 1e-07
UniRef100_C5XBT9 Putative uncharacterized protein Sb02g005130 n=... 61 1e-07
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 61 1e-07
UniRef100_A4RZU2 Predicted protein n=1 Tax=Ostreococcus lucimari... 61 1e-07
UniRef100_B4ND74 GK24962 n=1 Tax=Drosophila willistoni RepID=B4N... 61 1e-07
UniRef100_B4M7B5 GJ16487 n=1 Tax=Drosophila virilis RepID=B4M7B5... 61 1e-07
UniRef100_B4HH21 GM26598 n=1 Tax=Drosophila sechellia RepID=B4HH... 61 1e-07
UniRef100_Q9QYX7-2 Isoform 2 of Protein piccolo n=1 Tax=Mus musc... 61 1e-07
UniRef100_Q9QYX7-3 Isoform 3 of Protein piccolo n=1 Tax=Mus musc... 61 1e-07
UniRef100_Q9QYX7-4 Isoform 4 of Protein piccolo n=1 Tax=Mus musc... 61 1e-07
UniRef100_Q9QYX7 Protein piccolo n=1 Tax=Mus musculus RepID=PCLO... 61 1e-07
UniRef100_UPI0001862007 hypothetical protein BRAFLDRAFT_58365 n=... 61 1e-07
UniRef100_UPI0000DA32FB PREDICTED: hypothetical protein n=1 Tax=... 61 1e-07
UniRef100_Q94C69 Putative glycine-rich, zinc-finger DNA-binding ... 61 1e-07
UniRef100_Q966I7 Putative uncharacterized protein n=1 Tax=Caenor... 61 1e-07
UniRef100_Q86EQ4 Clone ZZD1536 mRNA sequence n=1 Tax=Schistosoma... 61 1e-07
UniRef100_Q5DE60 SJCHGC02361 protein (Fragment) n=1 Tax=Schistos... 61 1e-07
UniRef100_C3ZP49 Putative uncharacterized protein n=1 Tax=Branch... 61 1e-07
UniRef100_A7SMB3 Predicted protein n=1 Tax=Nematostella vectensi... 61 1e-07
UniRef100_UPI0000D9F56F PREDICTED: similar to Protein CXorf45 n=... 61 2e-07
UniRef100_UPI0000D9E82E PREDICTED: similar to additional sex com... 61 2e-07
UniRef100_UPI0000D9E82D PREDICTED: similar to additional sex com... 61 2e-07
UniRef100_UPI0001A2D80C UPI0001A2D80C related cluster n=1 Tax=Da... 61 2e-07
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 61 2e-07
UniRef100_B6IMJ6 Putative uncharacterized protein n=1 Tax=Rhodos... 61 2e-07
UniRef100_B4U926 Putative uncharacterized protein n=1 Tax=Hydrog... 61 2e-07
UniRef100_A3WBS5 Peptidoglycan-associated protein n=1 Tax=Erythr... 61 2e-07
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 61 2e-07
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 61 2e-07
UniRef100_Q4DD51 Putative uncharacterized protein n=1 Tax=Trypan... 61 2e-07
UniRef100_B7PLD9 Circumsporozoite protein, putative (Fragment) n... 61 2e-07
UniRef100_Q96VI4 Protease 1 n=1 Tax=Pneumocystis carinii RepID=Q... 61 2e-07
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 61 2e-07
UniRef100_UPI0000DC1359 UPI0000DC1359 related cluster n=1 Tax=Ra... 61 2e-07
UniRef100_A9EYW0 RNA-binding region RNP-1 (RNA recognition motif... 61 2e-07
UniRef100_A6G232 RNA-binding protein n=1 Tax=Plesiocystis pacifi... 61 2e-07
UniRef100_A6G1X8 Single-stranded DNA-binding protein n=1 Tax=Ple... 61 2e-07
UniRef100_A4CWS8 RNA-binding region RNP-1 (RNA recognition motif... 61 2e-07
UniRef100_P93517 Putative glycine-rich protein protein (Fragment... 61 2e-07
UniRef100_C5XP48 Putative uncharacterized protein Sb03g005056 (F... 61 2e-07
UniRef100_C1E5B0 Predicted protein n=1 Tax=Micromonas sp. RCC299... 61 2e-07
UniRef100_A9NNN7 Putative uncharacterized protein n=1 Tax=Picea ... 61 2e-07
UniRef100_B9PV72 Putative uncharacterized protein n=1 Tax=Toxopl... 61 2e-07
UniRef100_UPI0000DA3EC9 PREDICTED: similar to tumor endothelial ... 60 2e-07
UniRef100_UPI0000DA3E0C PREDICTED: similar to tumor endothelial ... 60 2e-07
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 60 2e-07
UniRef100_A4TTR4 Putative uncharacterized protein n=1 Tax=Magnet... 60 2e-07
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 60 2e-07
UniRef100_Q08195 Cysteine-rich extensin-like protein-2 n=1 Tax=N... 60 2e-07
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 60 2e-07
UniRef100_B9RS81 Serine-threonine protein kinase, plant-type, pu... 60 2e-07
UniRef100_B9GW18 Predicted protein n=1 Tax=Populus trichocarpa R... 60 2e-07
UniRef100_A4S9A6 Predicted protein n=1 Tax=Ostreococcus lucimari... 60 2e-07
UniRef100_B3M2B3 GF17078 n=1 Tax=Drosophila ananassae RepID=B3M2... 60 2e-07
UniRef100_B7Z9A8 cDNA FLJ58392 n=1 Tax=Homo sapiens RepID=B7Z9A8... 60 2e-07
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 60 2e-07
UniRef100_UPI0000DD90BE Os04g0438100 n=1 Tax=Oryza sativa Japoni... 60 2e-07
UniRef100_UPI000022125F Hypothetical protein CBG12875 n=1 Tax=Ca... 60 2e-07
UniRef100_Q8RW11 Putative glycine rich protein n=1 Tax=Rumex obt... 60 2e-07
UniRef100_C9SP10 ATP-dependent RNA helicase DBP2 n=1 Tax=Vertici... 60 2e-07
UniRef100_C9S6J1 Cellular nucleic acid-binding protein n=1 Tax=V... 60 2e-07
UniRef100_P49310 Glycine-rich RNA-binding protein GRP1A n=1 Tax=... 60 2e-07
UniRef100_Q05966 Glycine-rich RNA-binding protein 10 n=1 Tax=Bra... 60 2e-07
UniRef100_B5XQY2 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 60 2e-07
UniRef100_UPI00017974C7 PREDICTED: similar to KIAA1902 protein n... 60 3e-07
UniRef100_UPI0001795F40 PREDICTED: leiomodin 2 (cardiac) n=1 Tax... 60 3e-07
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 60 3e-07
UniRef100_UPI0000E1F764 PREDICTED: formin-like 2 isoform 6 n=1 T... 60 3e-07
UniRef100_UPI0000ECD10B Zinc finger homeobox protein 4 (Zinc fin... 60 3e-07
UniRef100_C3PP34 Actin polymerization protein RickA n=1 Tax=Rick... 60 3e-07
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 60 3e-07
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 60 3e-07
UniRef100_A1TG37 Putative uncharacterized protein n=1 Tax=Mycoba... 60 3e-07
UniRef100_C5JBL9 Uncharacterized protein n=1 Tax=uncultured bact... 60 3e-07
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 60 3e-07
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 60 3e-07
UniRef100_Q013S1 Chromosome 08 contig 1, DNA sequence n=1 Tax=Os... 60 3e-07
UniRef100_C5XK05 Putative uncharacterized protein Sb03g034303 (F... 60 3e-07
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 60 3e-07
UniRef100_A3B8S8 Putative uncharacterized protein n=1 Tax=Oryza ... 60 3e-07
UniRef100_B4NYJ9 GE18286 n=1 Tax=Drosophila yakuba RepID=B4NYJ9_... 60 3e-07
UniRef100_B3N3R7 GG25000 n=1 Tax=Drosophila erecta RepID=B3N3R7_... 60 3e-07
UniRef100_B0DQ50 Hypothetical proline-rich protein n=1 Tax=Lacca... 60 3e-07
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 60 3e-07
UniRef100_UPI000186773D hypothetical protein BRAFLDRAFT_237208 n... 60 3e-07
UniRef100_O65514 Putative glycine-rich cell wall protein n=1 Tax... 60 3e-07
UniRef100_O23646 RSZp22 protein n=1 Tax=Arabidopsis thaliana Rep... 60 3e-07
UniRef100_C5Z1R7 Putative uncharacterized protein Sb10g012740 n=... 60 3e-07
UniRef100_B7SFY1 Vasa n=1 Tax=Parhyale hawaiensis RepID=B7SFY1_9... 60 3e-07
UniRef100_C0SCG6 Cellular nucleic acid-binding protein n=2 Tax=P... 60 3e-07
UniRef100_UPI00017F09BE PREDICTED: similar to KIAA1902 protein, ... 60 4e-07
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 60 4e-07
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 60 4e-07
UniRef100_UPI0001951250 Formin-like protein 2 (Formin homology 2... 60 4e-07
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 60 4e-07
UniRef100_Q8YQB7 All3916 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 60 4e-07
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 60 4e-07
UniRef100_Q00TR5 Homology to unknown gene n=1 Tax=Ostreococcus t... 60 4e-07
UniRef100_Q00S27 Chromosome 19 contig 1, DNA sequence n=1 Tax=Os... 60 4e-07
UniRef100_C1E983 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 4e-07
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 60 4e-07
UniRef100_A7XQ02 Latex protein n=1 Tax=Morus alba RepID=A7XQ02_M... 60 4e-07
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 60 4e-07
UniRef100_B6AF23 Putative uncharacterized protein n=1 Tax=Crypto... 60 4e-07
UniRef100_B4P1N6 GE19123 (Fragment) n=1 Tax=Drosophila yakuba Re... 60 4e-07
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 60 4e-07
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 60 4e-07
UniRef100_C9S8M2 Cytokinesis protein sepA n=1 Tax=Verticillium a... 60 4e-07
UniRef100_C5MBX8 Putative uncharacterized protein n=1 Tax=Candid... 60 4e-07
UniRef100_B2AKS1 Translation initiation factor IF-2 n=1 Tax=Podo... 60 4e-07
UniRef100_P33485 Probable nuclear antigen n=2 Tax=Suid herpesvir... 60 4e-07
UniRef100_Q197B3 Putative membrane protein 047R n=1 Tax=Inverteb... 60 4e-07
UniRef100_Q9C6S1 Formin-like protein 14 n=1 Tax=Arabidopsis thal... 60 4e-07
UniRef100_P08001 Acrosin heavy chain n=1 Tax=Sus scrofa RepID=AC... 60 4e-07
UniRef100_P10323 Acrosin heavy chain n=1 Tax=Homo sapiens RepID=... 60 4e-07
UniRef100_UPI00019829E9 PREDICTED: hypothetical protein n=1 Tax=... 60 4e-07
UniRef100_UPI000180C465 PREDICTED: similar to cold-inducible RNA... 60 4e-07
UniRef100_Q7V9D6 RNA-binding region RNP-1 (RNA recognition motif... 60 4e-07
UniRef100_Q5YJM0 Glycine-rich protein (Fragment) n=1 Tax=Hyacint... 60 4e-07
UniRef100_C6JRV2 Putative uncharacterized protein Sb0012s004320 ... 60 4e-07
UniRef100_B3SW92 Glycine-rich protein 1 n=1 Tax=Boea hygrometric... 60 4e-07
UniRef100_A8HQM6 Predicted protein n=1 Tax=Chlamydomonas reinhar... 60 4e-07
UniRef100_A5BHG0 Putative uncharacterized protein n=1 Tax=Vitis ... 60 4e-07
UniRef100_B7PCR0 Cement protein, putative (Fragment) n=1 Tax=Ixo... 60 4e-07
UniRef100_B4X8C4 Vasa-like protein n=1 Tax=Hydractinia echinata ... 60 4e-07
UniRef100_B4IZJ4 GH14508 n=1 Tax=Drosophila grimshawi RepID=B4IZ... 60 4e-07
UniRef100_A7UTZ4 AGAP005965-PA (Fragment) n=1 Tax=Anopheles gamb... 60 4e-07
UniRef100_Q2GSQ8 Putative uncharacterized protein n=1 Tax=Chaeto... 60 4e-07
UniRef100_Q8T8R1 CCHC-type zinc finger protein CG3800 n=1 Tax=Dr... 60 4e-07
UniRef100_UPI0001B58635 hypothetical protein StAA4_25414 n=1 Tax... 59 5e-07
UniRef100_UPI0001923DA0 PREDICTED: similar to predicted protein ... 59 5e-07
UniRef100_UPI000186A01D hypothetical protein BRAFLDRAFT_106142 n... 59 5e-07
UniRef100_UPI000176023F PREDICTED: similar to protein kinase bet... 59 5e-07
UniRef100_UPI0000F2E472 PREDICTED: hypothetical protein n=1 Tax=... 59 5e-07
UniRef100_UPI000069FBE3 Formin-like protein 2 (Formin homology 2... 59 5e-07
UniRef100_UPI00016E10B6 UPI00016E10B6 related cluster n=1 Tax=Ta... 59 5e-07
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 59 5e-07
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 59 5e-07
UniRef100_UPI00016E10A1 UPI00016E10A1 related cluster n=1 Tax=Ta... 59 5e-07
UniRef100_UPI0001951234 Ras-associated and pleckstrin homology d... 59 5e-07
UniRef100_B4RDQ2 OmpA family protein n=1 Tax=Phenylobacterium zu... 59 5e-07
UniRef100_B4YB55 BimA n=1 Tax=Burkholderia pseudomallei RepID=B4... 59 5e-07
UniRef100_Q7XRW4 OSJNBb0062H02.5 protein n=2 Tax=Oryza sativa Re... 59 5e-07
UniRef100_Q013M1 Chromosome 08 contig 1, DNA sequence n=1 Tax=Os... 59 5e-07
UniRef100_C1MSQ5 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 5e-07
UniRef100_C1E755 Putative uncharacterized protein n=1 Tax=Microm... 59 5e-07
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 59 5e-07
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 59 5e-07
UniRef100_Q7YY78 Protease, possible n=2 Tax=Cryptosporidium parv... 59 5e-07
UniRef100_B4LT77 GJ19953 n=1 Tax=Drosophila virilis RepID=B4LT77... 59 5e-07
UniRef100_A8P9A8 Putative uncharacterized protein n=1 Tax=Brugia... 59 5e-07
UniRef100_A7SGL4 Predicted protein n=1 Tax=Nematostella vectensi... 59 5e-07
UniRef100_Q4WDJ2 Cytokinesis protein SepA/Bni1 n=1 Tax=Aspergill... 59 5e-07
UniRef100_C6H7E8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 59 5e-07
UniRef100_UPI0000122A7F Hypothetical protein CBG13587 n=1 Tax=Ca... 59 5e-07