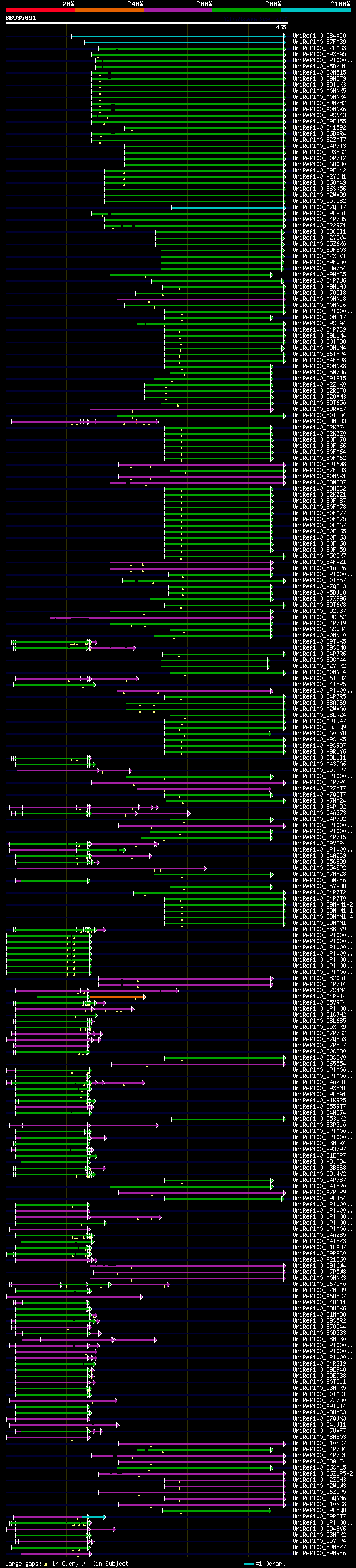
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB935691 RCC08518
(465 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q84XC0 Calcineurin B-like-interacting protein kinase n... 177 2e-43
UniRef100_B7FM39 Putative uncharacterized protein n=1 Tax=Medica... 168 1e-40
UniRef100_Q2LAG3 Protein kinase SRK n=1 Tax=Nicotiana tabacum Re... 122 9e-27
UniRef100_B9S8A5 CBL-interacting serine/threonine-protein kinase... 119 8e-26
UniRef100_UPI0001984050 PREDICTED: hypothetical protein n=1 Tax=... 119 1e-25
UniRef100_A5BKH1 CBL-interacting protein kinase 01 n=1 Tax=Vitis... 119 1e-25
UniRef100_C0M515 CBL-interacting protein kinase 19 n=1 Tax=Popul... 117 3e-25
UniRef100_B9NIF9 Predicted protein (Fragment) n=1 Tax=Populus tr... 116 7e-25
UniRef100_B9I1K3 Predicted protein (Fragment) n=1 Tax=Populus tr... 116 7e-25
UniRef100_A0MNK5 CBL-interacting protein kinase 18 n=1 Tax=Popul... 116 7e-25
UniRef100_A0MNK4 CBL-interacting protein kinase 17 n=1 Tax=Popul... 116 7e-25
UniRef100_B9H2H2 Predicted protein n=1 Tax=Populus trichocarpa R... 116 9e-25
UniRef100_A0MNK6 CBL-interacting protein kinase 19 n=1 Tax=Popul... 116 9e-25
UniRef100_Q9SN43 CBL-interacting serine/threonine-protein kinase... 113 7e-24
UniRef100_Q9FJ55 CBL-interacting serine/threonine-protein kinase... 112 1e-23
UniRef100_Q41592 Wpk4 protein kinase n=1 Tax=Triticum aestivum R... 107 3e-22
UniRef100_Q6DXR4 Putative serine-threonine kinase n=1 Tax=Gossyp... 107 4e-22
UniRef100_B2ZAT7 Putative serine-threonine kinase n=1 Tax=Gossyp... 107 5e-22
UniRef100_C4P7T3 CBL-interacting protein kinase 12 n=1 Tax=Sorgh... 104 3e-21
UniRef100_Q9SEG2 Protein kinase PK4 n=1 Tax=Zea mays RepID=Q9SEG... 103 6e-21
UniRef100_C0P7I2 Putative uncharacterized protein n=1 Tax=Zea ma... 103 6e-21
UniRef100_B6U0U0 CBL-interacting serine/threonine-protein kinase... 103 6e-21
UniRef100_B9FL42 Putative uncharacterized protein n=1 Tax=Oryza ... 102 2e-20
UniRef100_A2Y6H1 Putative uncharacterized protein n=1 Tax=Oryza ... 102 2e-20
UniRef100_Q68Y49 CBL-interacting protein kinase 19 n=2 Tax=Oryza... 102 2e-20
UniRef100_B6SK56 CBL-interacting serine/threonine-protein kinase... 100 5e-20
UniRef100_A2WV99 Putative uncharacterized protein n=1 Tax=Oryza ... 100 5e-20
UniRef100_Q5JLS2 CBL-interacting protein kinase 12 n=3 Tax=Oryza... 100 5e-20
UniRef100_A7QDI7 Chromosome chr10 scaffold_81, whole genome shot... 100 7e-20
UniRef100_Q9LP51 CBL-interacting serine/threonine-protein kinase... 99 1e-19
UniRef100_C4P7U5 CBL-interacting protein kinase 13 n=1 Tax=Sorgh... 99 1e-19
UniRef100_O22971 CBL-interacting serine/threonine-protein kinase... 92 2e-17
UniRef100_C8CBI1 CBL-interacting protein kinase 25 n=1 Tax=Oryza... 91 4e-17
UniRef100_A2YDV4 Putative uncharacterized protein n=1 Tax=Oryza ... 91 4e-17
UniRef100_Q5Z6X0 CBL-interacting protein kinase 25 n=2 Tax=Oryza... 91 4e-17
UniRef100_B9FE03 Putative uncharacterized protein n=1 Tax=Oryza ... 89 2e-16
UniRef100_A2XQV1 Putative uncharacterized protein n=1 Tax=Oryza ... 89 2e-16
UniRef100_B9EW50 Putative uncharacterized protein n=1 Tax=Oryza ... 84 6e-15
UniRef100_B8A754 Putative uncharacterized protein n=1 Tax=Oryza ... 81 3e-14
UniRef100_A9NXS5 Putative uncharacterized protein n=1 Tax=Picea ... 79 2e-13
UniRef100_C4P7U6 CBL-interacting protein kinase 32 n=1 Tax=Sorgh... 78 3e-13
UniRef100_A9NWA3 Putative uncharacterized protein n=1 Tax=Picea ... 77 5e-13
UniRef100_A7QDI8 CBL-interacting protein kinase 16 n=1 Tax=Vitis... 73 1e-11
UniRef100_A0MNJ8 CBL-interacting protein kinase 11 n=1 Tax=Popul... 73 1e-11
UniRef100_A0MNJ6 CBL-interacting protein kinase 9 n=2 Tax=Populu... 72 2e-11
UniRef100_UPI0001984051 PREDICTED: hypothetical protein isoform ... 71 3e-11
UniRef100_C0M517 CBL-interacting protein kinase 22 n=1 Tax=Popul... 70 6e-11
UniRef100_B9S8A4 CBL-interacting serine/threonine-protein kinase... 70 7e-11
UniRef100_C4P7S9 CBL-interacting protein kinase 20 n=1 Tax=Sorgh... 70 9e-11
UniRef100_Q9LWM4 CBL-interacting protein kinase 5 n=3 Tax=Oryza ... 70 9e-11
UniRef100_C0IRD0 CBL-interacting protein kinase 6 n=1 Tax=Cicer ... 69 1e-10
UniRef100_A9NWN4 Putative uncharacterized protein n=1 Tax=Picea ... 69 1e-10
UniRef100_B6THP4 CBL-interacting serine/threonine-protein kinase... 69 2e-10
UniRef100_B4F898 Putative uncharacterized protein n=1 Tax=Zea ma... 69 2e-10
UniRef100_A0MNK8 CBL-interacting protein kinase 22 n=2 Tax=Popul... 69 2e-10
UniRef100_Q5W736 CBL-interacting protein kinase 18 n=4 Tax=Oryza... 68 3e-10
UniRef100_B9IPI5 Predicted protein n=1 Tax=Populus trichocarpa R... 68 4e-10
UniRef100_A2ZHK0 Putative uncharacterized protein n=1 Tax=Oryza ... 67 5e-10
UniRef100_Q2RBF0 CBL-interacting protein kinase 15 n=2 Tax=Oryza... 67 5e-10
UniRef100_Q2QYM3 CBL-interacting protein kinase 14 n=2 Tax=Oryza... 67 5e-10
UniRef100_B9T650 CBL-interacting serine/threonine-protein kinase... 67 8e-10
UniRef100_B9RVE7 CBL-interacting serine/threonine-protein kinase... 67 8e-10
UniRef100_B0I554 Protein kinase n=1 Tax=Phaseolus vulgaris RepID... 67 8e-10
UniRef100_B3M2B3 GF17078 n=1 Tax=Drosophila ananassae RepID=B3M2... 52 1e-09
UniRef100_B2KZZ4 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B2KZZ0 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B0FM70 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B0FM66 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B0FM64 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B0FM62 Serine-threonine kinase n=1 Tax=Persea american... 66 1e-09
UniRef100_B9I6W8 Predicted protein n=1 Tax=Populus trichocarpa R... 66 1e-09
UniRef100_B7FIU3 Putative uncharacterized protein n=1 Tax=Medica... 66 1e-09
UniRef100_A0MNK1 CBL-interacting protein kinase 14 n=1 Tax=Popul... 66 1e-09
UniRef100_Q8W2D7 CBL-interacting protein kinase n=1 Tax=Brassica... 65 2e-09
UniRef100_Q8H2C2 Serine/threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B2KZZ1 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM87 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM78 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM77 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM75 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM67 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM65 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM63 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM60 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_B0FM59 Serine-threonine kinase n=1 Tax=Persea american... 65 2e-09
UniRef100_A5C5K7 CBL-interacting protein kinase 03 n=1 Tax=Vitis... 65 2e-09
UniRef100_B4FXZ1 CBL-interacting serine/threonine-protein kinase... 65 2e-09
UniRef100_B1A5P6 CBL-interacting protein kinase n=1 Tax=Zea mays... 65 2e-09
UniRef100_UPI0001983BB5 PREDICTED: hypothetical protein isoform ... 65 3e-09
UniRef100_B0I557 Protein kinase n=1 Tax=Phaseolus vulgaris RepID... 65 3e-09
UniRef100_A7QFL3 Chromosome chr8 scaffold_88, whole genome shotg... 65 3e-09
UniRef100_A5BJJ8 CBL-interacting protein kinase 07 n=1 Tax=Vitis... 65 3e-09
UniRef100_Q7X996 CBL-interacting protein kinase 2 n=3 Tax=Oryza ... 65 3e-09
UniRef100_B9T6V8 CBL-interacting serine/threonine-protein kinase... 64 4e-09
UniRef100_P92937 CBL-interacting serine/threonine-protein kinase... 64 4e-09
UniRef100_Q9C562 CBL-interacting serine/threonine-protein kinase... 64 4e-09
UniRef100_C4P7T9 CBL-interacting protein kinase 02 n=1 Tax=Sorgh... 64 5e-09
UniRef100_B6SW34 CBL-interacting serine/threonine-protein kinase... 64 5e-09
UniRef100_A0MNJ0 CBL-interacting protein kinase 2 n=1 Tax=Populu... 64 5e-09
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 64 7e-09
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 64 7e-09
UniRef100_C4P7R6 CBL-interacting protein kinase 04 n=1 Tax=Selag... 64 7e-09
UniRef100_B9G044 Putative uncharacterized protein n=1 Tax=Oryza ... 64 7e-09
UniRef100_A2YTK2 Putative uncharacterized protein n=1 Tax=Oryza ... 64 7e-09
UniRef100_A0MNJ4 CBL-interacting protein kinase 7 n=1 Tax=Populu... 63 9e-09
UniRef100_C6TLD2 Putative uncharacterized protein n=1 Tax=Glycin... 46 1e-08
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 63 1e-08
UniRef100_UPI0001983733 PREDICTED: hypothetical protein isoform ... 63 1e-08
UniRef100_C4P7R5 CBL-interacting protein kinase 03 n=1 Tax=Selag... 63 1e-08
UniRef100_B8A9S9 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_A2WVA0 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_Q8LK24 SOS2-like protein kinase n=1 Tax=Glycine max Re... 62 2e-08
UniRef100_A9T947 Predicted protein n=2 Tax=Physcomitrella patens... 62 2e-08
UniRef100_Q5JLQ9 CBL-interacting protein kinase 30 n=2 Tax=Oryza... 62 2e-08
UniRef100_Q60EY8 CBL-interacting protein kinase 20 n=3 Tax=Oryza... 62 2e-08
UniRef100_A9SHK5 Predicted protein n=2 Tax=Physcomitrella patens... 62 2e-08
UniRef100_A9S987 Predicted protein n=2 Tax=Physcomitrella patens... 62 2e-08
UniRef100_A9RUY6 Predicted protein n=2 Tax=Physcomitrella patens... 62 2e-08
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 62 3e-08
UniRef100_A4S9A6 Predicted protein n=1 Tax=Ostreococcus lucimari... 62 3e-08
UniRef100_C5JPP7 Predicted protein n=1 Tax=Ajellomyces dermatiti... 62 3e-08
UniRef100_UPI0001983732 PREDICTED: hypothetical protein isoform ... 62 3e-08
UniRef100_C4P7R4 CBL-interacting protein kinase 02 n=1 Tax=Selag... 62 3e-08
UniRef100_B2ZYT7 CBL-interacting protein kinase 1 n=1 Tax=Vicia ... 62 3e-08
UniRef100_A7Q3T7 CBL-interacting protein kinase 13 n=1 Tax=Vitis... 62 3e-08
UniRef100_A7NY24 CBL-interacting protein kinase 10 n=1 Tax=Vitis... 62 3e-08
UniRef100_B4PM92 GE24601 n=1 Tax=Drosophila yakuba RepID=B4PM92_... 52 3e-08
UniRef100_Q4A373 Putative lectin protein n=2 Tax=Emiliania huxle... 61 3e-08
UniRef100_C4P7U2 CBL-interacting protein kinase 15 n=1 Tax=Sorgh... 61 3e-08
UniRef100_UPI0001984DB1 PREDICTED: hypothetical protein n=1 Tax=... 61 4e-08
UniRef100_UPI0001983731 PREDICTED: hypothetical protein isoform ... 61 4e-08
UniRef100_C4P7T5 CBL-interacting protein kinase 30 n=1 Tax=Sorgh... 61 4e-08
UniRef100_Q9VEP4 CG5225 n=2 Tax=Drosophila melanogaster RepID=Q9... 52 5e-08
UniRef100_UPI0000F2CDB8 PREDICTED: similar to engrailed homolog ... 43 5e-08
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 60 6e-08
UniRef100_C5G899 Predicted protein n=1 Tax=Ajellomyces dermatiti... 60 6e-08
UniRef100_Q54SP2 Formin-B n=1 Tax=Dictyostelium discoideum RepID... 60 6e-08
UniRef100_A7NY28 CBL-interacting protein kinase 11 n=1 Tax=Vitis... 60 6e-08
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 60 7e-08
UniRef100_C5YVU8 Putative uncharacterized protein Sb09g013590 n=... 60 7e-08
UniRef100_C4P7T2 CBL-interacting protein kinase 06 n=1 Tax=Sorgh... 60 7e-08
UniRef100_C4P7T0 CBL-interacting protein kinase 18 n=1 Tax=Sorgh... 60 7e-08
UniRef100_Q9MAM1-2 Isoform 2 of CBL-interacting serine/threonine... 60 7e-08
UniRef100_Q9MAM1-1 Isoform 1 of CBL-interacting serine/threonine... 60 7e-08
UniRef100_Q9MAM1-4 Isoform 4 of CBL-interacting serine/threonine... 60 7e-08
UniRef100_Q9MAM1 CBL-interacting serine/threonine-protein kinase... 60 7e-08
UniRef100_B8BCY9 Putative uncharacterized protein n=1 Tax=Oryza ... 58 9e-08
UniRef100_UPI00016E59B7 UPI00016E59B7 related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E59B6 UPI00016E59B6 related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E599B UPI00016E599B related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E599A UPI00016E599A related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E5999 UPI00016E5999 related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E597F UPI00016E597F related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E595C UPI00016E595C related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_O82051 Putative serine/threonine protein kinase n=1 Ta... 60 1e-07
UniRef100_C4P7T4 CBL-interacting protein kinase 31 n=1 Tax=Sorgh... 60 1e-07
UniRef100_Q7S4M4 Predicted protein n=1 Tax=Neurospora crassa Rep... 41 1e-07
UniRef100_B4PA14 GE12142 n=1 Tax=Drosophila yakuba RepID=B4PA14_... 50 1e-07
UniRef100_Q5VRF4 Os06g0168700 protein n=1 Tax=Oryza sativa Japon... 58 1e-07
UniRef100_UPI0001982977 PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_Q1G7H2 Dishevelled-associated activator of morphogenes... 59 1e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 59 1e-07
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 59 1e-07
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 59 1e-07
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 59 1e-07
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 59 1e-07
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 59 1e-07
UniRef100_Q8S3V0 Putative serine threonine kinase n=1 Tax=Sander... 59 1e-07
UniRef100_O65554 CBL-interacting serine/threonine-protein kinase... 59 1e-07
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 59 2e-07
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 59 2e-07
UniRef100_Q9FXA1 AT1G49750 protein n=1 Tax=Arabidopsis thaliana ... 59 2e-07
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 59 2e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 59 2e-07
UniRef100_B4ND74 GK24962 n=1 Tax=Drosophila willistoni RepID=B4N... 59 2e-07
UniRef100_Q53UK2 Ser/Thr protein kinase (Fragment) n=1 Tax=Lotus... 59 2e-07
UniRef100_B3P3J0 GG21917 n=1 Tax=Drosophila erecta RepID=B3P3J0_... 54 2e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 59 2e-07
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 59 2e-07
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 59 2e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 59 2e-07
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 59 2e-07
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 59 2e-07
UniRef100_A3B8S8 Putative uncharacterized protein n=1 Tax=Oryza ... 59 2e-07
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 59 2e-07
UniRef100_C4P7S7 CBL-interacting protein kinase 25 n=1 Tax=Sorgh... 59 2e-07
UniRef100_C4IYR0 Putative uncharacterized protein n=1 Tax=Zea ma... 59 2e-07
UniRef100_A7PXR9 CBL-interacting protein kinase 12 n=1 Tax=Vitis... 59 2e-07
UniRef100_Q9FJ54 CBL-interacting serine/threonine-protein kinase... 59 2e-07
UniRef100_UPI000198409E PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 58 3e-07
UniRef100_UPI00017EFCA0 PREDICTED: dishevelled associated activa... 58 3e-07
UniRef100_UPI0000E47472 PREDICTED: similar to Splicing factor 3b... 58 3e-07
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 58 3e-07
UniRef100_A4TEZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 58 3e-07
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 58 3e-07
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 58 3e-07
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 58 3e-07
UniRef100_B9I6W4 Predicted protein n=1 Tax=Populus trichocarpa R... 58 3e-07
UniRef100_A7P5W8 CBL-interacting protein kinase 15 n=1 Tax=Vitis... 58 3e-07
UniRef100_A0MNK3 CBL-interacting protein kinase 16 n=1 Tax=Popul... 58 3e-07
UniRef100_Q67WF0 Zinc finger (CCCH-type) protein-like n=1 Tax=Or... 46 3e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 58 4e-07
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 58 4e-07
UniRef100_C4B111 Intracellular motility protein A n=4 Tax=Burkho... 58 4e-07
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 58 4e-07
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 58 4e-07
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 58 4e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 58 4e-07
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 58 4e-07
UniRef100_Q8MP30 Uncharacterized histidine-rich protein DDB01677... 58 4e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 57 5e-07
UniRef100_UPI000155C36A PREDICTED: hypothetical protein, partial... 57 5e-07
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 57 5e-07
UniRef100_Q4RSI9 Chromosome 13 SCAF15000, whole genome shotgun s... 57 5e-07
UniRef100_Q9E940 ICP4 protein n=1 Tax=Gallid herpesvirus 3 RepID... 57 5e-07
UniRef100_Q9E938 ICP4 protein n=1 Tax=Gallid herpesvirus 3 RepID... 57 5e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 57 5e-07
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 57 5e-07
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 57 5e-07
UniRef100_C7J750 Os09g0496100 protein (Fragment) n=1 Tax=Oryza s... 57 5e-07
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 57 5e-07
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 57 5e-07
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 57 5e-07
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 57 5e-07
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 57 5e-07
UniRef100_A8NE03 Predicted protein n=1 Tax=Coprinopsis cinerea o... 57 5e-07
UniRef100_Q10SC7 CIPK-like protein 1, putative, expressed n=1 Ta... 57 5e-07
UniRef100_C4P7U4 CBL-interacting protein kinase 10 (Fragment) n=... 57 5e-07
UniRef100_C4P7S1 CBL-interacting protein kinase 09 n=1 Tax=Sorgh... 57 5e-07
UniRef100_B8AMF4 Putative uncharacterized protein n=1 Tax=Oryza ... 57 5e-07
UniRef100_B6SXL5 CBL-interacting serine/threonine-protein kinase... 57 5e-07
UniRef100_Q6ZLP5-2 Isoform 2 of CBL-interacting protein kinase 2... 57 5e-07
UniRef100_A2ZQH3 Putative uncharacterized protein n=1 Tax=Oryza ... 57 5e-07
UniRef100_A2WLW3 Putative uncharacterized protein n=1 Tax=Oryza ... 57 5e-07
UniRef100_Q6ZLP5 CBL-interacting protein kinase 23 n=1 Tax=Oryza... 57 5e-07
UniRef100_Q5QNM6 Putative CBL-interacting protein kinase 13 n=1 ... 57 5e-07
UniRef100_Q10SC8 CBL-interacting protein kinase 9 n=2 Tax=Oryza ... 57 5e-07
UniRef100_Q9LYQ8 CBL-interacting serine/threonine-protein kinase... 57 5e-07
UniRef100_B9RTT7 Mahogunin, putative n=1 Tax=Ricinus communis Re... 43 5e-07
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 57 6e-07
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 57 6e-07
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 57 6e-07
UniRef100_B9N8Z7 Predicted protein (Fragment) n=2 Tax=Populus tr... 57 6e-07
UniRef100_B9H9E6 Predicted protein (Fragment) n=1 Tax=Populus tr... 57 6e-07
UniRef100_A7XQ02 Latex protein n=1 Tax=Morus alba RepID=A7XQ02_M... 57 6e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 57 6e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 57 6e-07
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 57 6e-07
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 57 6e-07
UniRef100_B9SMX5 CBL-interacting serine/threonine-protein kinase... 57 6e-07
UniRef100_A9RKW0 Predicted protein n=2 Tax=Physcomitrella patens... 57 6e-07
UniRef100_Q26056 Histidine-rich protein (Fragment) n=1 Tax=Plasm... 57 6e-07
UniRef100_P04929 Histidine-rich glycoprotein n=1 Tax=Plasmodium ... 57 6e-07
UniRef100_B3MA92 GF25137 n=1 Tax=Drosophila ananassae RepID=B3MA... 42 7e-07
UniRef100_B3MU38 GF21178 n=1 Tax=Drosophila ananassae RepID=B3MU... 40 7e-07
UniRef100_UPI0000F2B946 PREDICTED: similar to tumor endothelial ... 57 8e-07
UniRef100_Q4A263 Putative membrane protein n=1 Tax=Emiliania hux... 57 8e-07
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 57 8e-07
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 57 8e-07
UniRef100_Q5ZB68 Os01g0594300 protein n=1 Tax=Oryza sativa Japon... 57 8e-07
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 57 8e-07
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 57 8e-07
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 57 8e-07
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 57 8e-07
UniRef100_Q296I6 GA17823 n=1 Tax=Drosophila pseudoobscura pseudo... 57 8e-07
UniRef100_O01900 Putative uncharacterized protein n=1 Tax=Caenor... 57 8e-07
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_A3MS59 Putative uncharacterized protein n=1 Tax=Pyroba... 57 8e-07
UniRef100_C4P7S2 CBL-interacting protein kinase 23 n=1 Tax=Sorgh... 57 8e-07
UniRef100_C0P7R9 Putative uncharacterized protein n=2 Tax=Zea ma... 57 8e-07
UniRef100_C0M520 CBL-interacting protein kinase 25 n=1 Tax=Popul... 57 8e-07
UniRef100_B9T407 CBL-interacting serine/threonine-protein kinase... 57 8e-07
UniRef100_B9SVF5 ATP binding protein, putative n=1 Tax=Ricinus c... 57 8e-07
UniRef100_B8B7B8 Putative uncharacterized protein n=1 Tax=Oryza ... 57 8e-07
UniRef100_B4FU99 Putative uncharacterized protein n=1 Tax=Zea ma... 57 8e-07
UniRef100_A5HLX6 CBL-interacting protein kinase n=1 Tax=Hordeum ... 57 8e-07
UniRef100_UPI0000F22463 IQ motif and Sec7 domain 2 n=1 Tax=Mus m... 43 9e-07
UniRef100_A4GZ26 ARF6 guanine nucleotide exchange factor IQArfGE... 43 9e-07
UniRef100_UPI00015DF451 IQ motif and Sec7 domain 2 n=1 Tax=Mus m... 43 9e-07
UniRef100_UPI000179E2B1 UPI000179E2B1 related cluster n=1 Tax=Bo... 43 9e-07
UniRef100_UPI0001797DF3 PREDICTED: similar to IQ motif and Sec7 ... 43 9e-07
UniRef100_B4J560 GH20878 n=1 Tax=Drosophila grimshawi RepID=B4J5... 50 1e-06
UniRef100_Q54KT2 Putative uncharacterized protein DDB_G0287191 n... 47 1e-06
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 56 1e-06
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 56 1e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 56 1e-06
UniRef100_A5G2K8 Putative uncharacterized protein n=1 Tax=Acidip... 56 1e-06
UniRef100_C0UNU2 Uncharacterized stress response protein, TerZ-a... 56 1e-06
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 56 1e-06
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_Q7YY78 Protease, possible n=2 Tax=Cryptosporidium parv... 56 1e-06
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 56 1e-06
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B9EU80 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_B8ABQ0 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_B6SH63 CBL-interacting serine/threonine-protein kinase... 56 1e-06
UniRef100_A5BQ82 CBL-interacting protein kinase 04 n=1 Tax=Vitis... 56 1e-06
UniRef100_Q0JI49 CBL-interacting protein kinase 11 n=2 Tax=Oryza... 56 1e-06
UniRef100_B5DWM1 GA26446 n=1 Tax=Drosophila pseudoobscura pseudo... 46 1e-06
UniRef100_C5Z6L1 Putative uncharacterized protein Sb10g024955 (F... 40 1e-06
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 56 1e-06
UniRef100_C9LW10 Outer membrane autotransporter barrel domain pr... 56 1e-06
UniRef100_Q948Y9 VMP1 protein n=1 Tax=Volvox carteri f. nagarien... 56 1e-06
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 56 1e-06
UniRef100_Q8S9B4 Matrix metalloproteinase n=1 Tax=Volvox carteri... 56 1e-06
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 56 1e-06
UniRef100_B9RYC5 Serine-threonine protein kinase, plant-type, pu... 56 1e-06
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 56 1e-06
UniRef100_A7USF3 AGAP000510-PA (Fragment) n=1 Tax=Anopheles gamb... 56 1e-06
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 56 1e-06
UniRef100_Q9XFJ3 Putative serine/threonine protein kinase n=1 Ta... 56 1e-06
UniRef100_B9T2W2 CBL-interacting serine/threonine-protein kinase... 56 1e-06
UniRef100_B9F8D1 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_B8APG6 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_A7Q176 Chromosome chr10 scaffold_43, whole genome shot... 56 1e-06
UniRef100_A5B4V8 CBL-interacting protein kinase 06 n=1 Tax=Vitis... 56 1e-06
UniRef100_A0MNL1 CBL-interacting protein kinase 25 n=2 Tax=Popul... 56 1e-06
UniRef100_Q10LQ2 CBL-interacting protein kinase 10 n=2 Tax=Oryza... 56 1e-06
UniRef100_B4LT77 GJ19953 n=1 Tax=Drosophila virilis RepID=B4LT77... 43 1e-06
UniRef100_UPI00017C2E80 PREDICTED: similar to forkhead box B2 n=... 42 1e-06
UniRef100_UPI0001861057 hypothetical protein BRAFLDRAFT_119668 n... 55 2e-06
UniRef100_UPI0000DA45C5 PREDICTED: similar to diacylglycerol kin... 55 2e-06
UniRef100_UPI0000DA19D8 PREDICTED: similar to MAGE-like protein ... 55 2e-06
UniRef100_UPI0000DC20B8 UPI0000DC20B8 related cluster n=1 Tax=Ra... 55 2e-06
UniRef100_Q8BEN8 ORF58 n=1 Tax=Callitrichine herpesvirus 3 RepID... 55 2e-06
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 55 2e-06
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 2e-06
UniRef100_Q9FLQ7 Formin-like protein 20 n=1 Tax=Arabidopsis thal... 55 2e-06
UniRef100_UPI000198442C PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI00019842D2 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_Q4QWQ4 Serine/threonine protein kinase (Fragment) n=1 ... 55 2e-06
UniRef100_C0P5L0 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_C0P3A2 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_B9N707 Predicted protein n=2 Tax=Populus trichocarpa R... 55 2e-06
UniRef100_A7Q6M5 CBL-interacting protein kinase 14 n=1 Tax=Vitis... 55 2e-06
UniRef100_A7BJ79 CBL-interacting protein kinase n=1 Tax=Vigna un... 55 2e-06
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI0001926BC7 PREDICTED: similar to mini-collagen isof... 55 2e-06
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 55 2e-06
UniRef100_Q5VR46 Os01g0180000 protein n=1 Tax=Oryza sativa Japon... 55 2e-06
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 55 2e-06
UniRef100_Q00YH8 LPA (ISS) n=1 Tax=Ostreococcus tauri RepID=Q00Y... 55 2e-06
UniRef100_O81765 Extensin-like protein n=1 Tax=Arabidopsis thali... 55 2e-06
UniRef100_B8ADK4 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_A3BNX1 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_C6HGV1 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 55 2e-06
UniRef100_C0NZQ8 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 55 2e-06
UniRef100_Q95JC9-2 Isoform 2 of Basic proline-rich protein n=1 T... 55 2e-06
UniRef100_Q95JC9-3 Isoform 3 of Basic proline-rich protein n=1 T... 55 2e-06
UniRef100_C0P6L2 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_A0MNK0 CBL-interacting protein kinase 13 n=1 Tax=Popul... 55 2e-06
UniRef100_A7QDU5 Chromosome chr4 scaffold_83, whole genome shotg... 42 2e-06
UniRef100_Q0PIW3 HyPRP1 n=1 Tax=Gossypium hirsutum RepID=Q0PIW3_... 46 3e-06
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI00016E324C UPI00016E324C related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 55 3e-06
UniRef100_UPI0000F310CB PREDICTED: Bos taurus similar to trithor... 55 3e-06
UniRef100_B9LBU3 Putative uncharacterized protein n=1 Tax=Chloro... 55 3e-06
UniRef100_A9WHI6 Putative uncharacterized protein n=1 Tax=Chloro... 55 3e-06
UniRef100_Q00TR5 Homology to unknown gene n=1 Tax=Ostreococcus t... 55 3e-06
UniRef100_C1MSQ5 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 3e-06
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 3e-06
UniRef100_C1EC31 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 3e-06
UniRef100_B9SHA4 ATP binding protein, putative n=1 Tax=Ricinus c... 55 3e-06
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 55 3e-06
UniRef100_A7PES6 Chromosome chr11 scaffold_13, whole genome shot... 55 3e-06
UniRef100_Q5BHU8 AT04667p n=1 Tax=Drosophila melanogaster RepID=... 55 3e-06
UniRef100_Q29QD2 AT18380p n=1 Tax=Drosophila melanogaster RepID=... 55 3e-06
UniRef100_Q16U10 Putative uncharacterized protein n=1 Tax=Aedes ... 55 3e-06
UniRef100_B4N9Z0 GK12228 n=1 Tax=Drosophila willistoni RepID=B4N... 55 3e-06
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 55 3e-06
UniRef100_C4P7R3 CBL-interacting protein kinase 01 n=1 Tax=Selag... 55 3e-06
UniRef100_B3NAT6 GG24446 n=1 Tax=Drosophila erecta RepID=B3NAT6_... 55 3e-06
UniRef100_UPI0001B58635 hypothetical protein StAA4_25414 n=1 Tax... 54 4e-06
UniRef100_UPI000194BE5E PREDICTED: enabled homolog n=1 Tax=Taeni... 54 4e-06
UniRef100_UPI0001926BAB PREDICTED: similar to mini-collagen n=1 ... 54 4e-06
UniRef100_UPI000023F096 hypothetical protein FG08656.1 n=1 Tax=G... 54 4e-06
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 54 4e-06
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 54 4e-06
UniRef100_A1UCC3 Putative uncharacterized protein n=2 Tax=Mycoba... 54 4e-06
UniRef100_Q01AC3 Meltrins, fertilins and related Zn-dependent me... 54 4e-06
UniRef100_Q00X46 Chromosome 13 contig 1, DNA sequence n=1 Tax=Os... 54 4e-06
UniRef100_Q00TD0 Chromosome 17 contig 1, DNA sequence. (Fragment... 54 4e-06
UniRef100_C1EC93 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 4e-06
UniRef100_B9RS81 Serine-threonine protein kinase, plant-type, pu... 54 4e-06
UniRef100_Q7PNI8 AGAP000892-PA (Fragment) n=1 Tax=Anopheles gamb... 54 4e-06
UniRef100_B4GF40 GL22134 n=1 Tax=Drosophila persimilis RepID=B4G... 54 4e-06
UniRef100_C0NGH8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 54 4e-06
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 54 4e-06
UniRef100_B0CT66 RhoA GTPase effector DIA/Diaphanous n=1 Tax=Lac... 54 4e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 54 4e-06
UniRef100_Q10M85 CIPK-like protein 1, putative, expressed n=1 Ta... 54 4e-06
UniRef100_O24342 Serine/threonine kinase n=1 Tax=Sorghum bicolor... 54 4e-06
UniRef100_C6TD54 Putative uncharacterized protein n=1 Tax=Glycin... 54 4e-06
UniRef100_C4P7T6 CBL-interacting protein kinase 03 n=1 Tax=Sorgh... 54 4e-06
UniRef100_B9T8H6 CBL-interacting serine/threonine-protein kinase... 54 4e-06
UniRef100_B8BIX7 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_B8AN11 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_Q10M84 CIPK-like protein 1, putative, expressed n=2 Ta... 54 4e-06
UniRef100_A2ZHU3 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_Q6X4A2-2 Isoform 2 of CBL-interacting protein kinase 3... 54 4e-06
UniRef100_Q6X4A2 CBL-interacting protein kinase 31 n=2 Tax=Oryza... 54 4e-06
UniRef100_B7Z017 CG33003, isoform B n=1 Tax=Drosophila melanogas... 44 4e-06
UniRef100_Q86BM9 CG33003, isoform A n=1 Tax=Drosophila melanogas... 44 4e-06
UniRef100_Q1A1A5 Forkhead box protein G1 n=1 Tax=Chlorocebus pyg... 39 4e-06
UniRef100_UPI0001862DED hypothetical protein BRAFLDRAFT_122351 n... 54 5e-06
UniRef100_UPI000179680F PREDICTED: similar to WW domain-binding ... 54 5e-06
UniRef100_UPI0000EBE793 PREDICTED: similar to MAGEL2 protein n=1... 54 5e-06
UniRef100_UPI0000D9B773 PREDICTED: similar to CG5514-PB, isoform... 54 5e-06
UniRef100_UPI00005A6092 PREDICTED: similar to multidomain presyn... 54 5e-06
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 54 5e-06
UniRef100_A3PW04 U5 snRNP spliceosome subunit-like protein n=1 T... 54 5e-06
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 54 5e-06
UniRef100_C1DYH3 Putative uncharacterized protein n=1 Tax=Microm... 54 5e-06
UniRef100_B9SMV7 Vegetative cell wall protein gp1, putative n=1 ... 54 5e-06
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 54 5e-06
UniRef100_B9EVM1 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 54 5e-06
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 54 5e-06
UniRef100_A2XD25 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_C3XX60 Putative uncharacterized protein n=1 Tax=Branch... 54 5e-06
UniRef100_B4MN04 GK17614 n=1 Tax=Drosophila willistoni RepID=B4M... 54 5e-06
UniRef100_A8QF93 EBNA-2 nuclear protein, putative n=1 Tax=Brugia... 54 5e-06
UniRef100_B8MHY0 Putative uncharacterized protein n=1 Tax=Talaro... 54 5e-06
UniRef100_B2VV45 Predicted protein n=1 Tax=Pyrenophora tritici-r... 54 5e-06
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 54 5e-06
UniRef100_Q3ULZ2 FH2 domain-containing protein 1 n=1 Tax=Mus mus... 54 5e-06
UniRef100_Q53VM3 Ser/Thr protein kinase n=1 Tax=Lotus japonicus ... 54 5e-06
UniRef100_C4P7S0 CBL-interacting protein kinase 27 n=1 Tax=Sorgh... 54 5e-06
UniRef100_B9S4C5 CBL-interacting serine/threonine-protein kinase... 54 5e-06
UniRef100_B8LQ83 Putative uncharacterized protein n=1 Tax=Picea ... 54 5e-06
UniRef100_B8A0K9 Putative uncharacterized protein n=1 Tax=Zea ma... 54 5e-06
UniRef100_B7U1M7 Cold-induced CIPK n=1 Tax=Lepidium latifolium R... 54 5e-06
UniRef100_B5AU21 CBL-interacting protein kinase n=1 Tax=Zea mays... 54 5e-06
UniRef100_A2X165 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_B4I052 GM12551 n=1 Tax=Drosophila sechellia RepID=B4I0... 54 5e-06
UniRef100_Q6H7U5 CBL-interacting protein kinase 26 n=2 Tax=Oryza... 54 5e-06
UniRef100_B4NYJ9 GE18286 n=1 Tax=Drosophila yakuba RepID=B4NYJ9_... 47 5e-06
UniRef100_UPI0000F2CD9F PREDICTED: similar to Splicing factor, a... 47 7e-06
UniRef100_B3NTG0 GG18108 n=1 Tax=Drosophila erecta RepID=B3NTG0_... 44 7e-06
UniRef100_A0NH94 AGAP001118-PA (Fragment) n=1 Tax=Anopheles gamb... 45 7e-06
UniRef100_B4GDZ6 GL21920 n=1 Tax=Drosophila persimilis RepID=B4G... 44 7e-06
UniRef100_UPI00019261C6 PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0000DC131C UPI0000DC131C related cluster n=1 Tax=Ra... 54 7e-06
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 54 7e-06
UniRef100_A4FC63 Putative uncharacterized protein n=1 Tax=Saccha... 54 7e-06
UniRef100_Q9XIB6 F13F21.7 protein n=1 Tax=Arabidopsis thaliana R... 54 7e-06
UniRef100_Q01DC8 Plg protein (ISS) n=1 Tax=Ostreococcus tauri Re... 54 7e-06
UniRef100_C1FHQ4 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 7e-06
UniRef100_C1E983 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 7e-06
UniRef100_B7PAV3 Putative uncharacterized protein (Fragment) n=1... 54 7e-06
UniRef100_B4I348 GM18116 n=1 Tax=Drosophila sechellia RepID=B4I3... 54 7e-06
UniRef100_A4H3P3 Putative uncharacterized protein n=1 Tax=Leishm... 54 7e-06
UniRef100_B4DMD3 cDNA FLJ58174, highly similar to WW domain-bind... 54 7e-06
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 54 7e-06
UniRef100_C1GXM9 Cytokinesis protein sepA n=1 Tax=Paracoccidioid... 54 7e-06
UniRef100_C0SGP4 Cytokinesis protein sepA n=1 Tax=Paracoccidioid... 54 7e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 54 7e-06
UniRef100_B0CS37 Predicted protein n=1 Tax=Laccaria bicolor S238... 54 7e-06
UniRef100_A8NHF1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 54 7e-06
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 54 7e-06
UniRef100_Q54WZ5 Probable inactive serine/threonine-protein kina... 54 7e-06
UniRef100_C4P7R9 CBL-interacting protein kinase 19 n=1 Tax=Sorgh... 54 7e-06
UniRef100_C0PK96 Putative uncharacterized protein n=1 Tax=Zea ma... 54 7e-06
UniRef100_Q2QY53 CBL-interacting protein kinase 32 n=1 Tax=Oryza... 54 7e-06
UniRef100_UPI0001760EE5 PREDICTED: similar to Casitas B-lineage ... 37 7e-06
UniRef100_UPI0001A2BE1D hypothetical protein LOC100002706 (LOC10... 37 7e-06
UniRef100_Q9ZWT0 Extensin n=1 Tax=Adiantum capillus-veneris RepI... 45 7e-06
UniRef100_P55316 Forkhead box protein G1 n=1 Tax=Homo sapiens Re... 38 9e-06
UniRef100_UPI000186A01D hypothetical protein BRAFLDRAFT_106142 n... 53 9e-06
UniRef100_UPI0001A2CA84 UPI0001A2CA84 related cluster n=1 Tax=Da... 53 9e-06
UniRef100_UPI00016EA046 UPI00016EA046 related cluster n=1 Tax=Ta... 53 9e-06
UniRef100_UPI0000ECC7C5 enabled homolog n=1 Tax=Gallus gallus Re... 53 9e-06
UniRef100_UPI0000ECC7C4 enabled homolog n=1 Tax=Gallus gallus Re... 53 9e-06
UniRef100_UPI0000ECC7C3 enabled homolog n=1 Tax=Gallus gallus Re... 53 9e-06
UniRef100_UPI00003ACBF3 enabled homolog n=1 Tax=Gallus gallus Re... 53 9e-06
UniRef100_Q9DEG2 AvenaII n=1 Tax=Gallus gallus RepID=Q9DEG2_CHICK 53 9e-06
UniRef100_Q9DEG1 AvenaIII n=1 Tax=Gallus gallus RepID=Q9DEG1_CHICK 53 9e-06
UniRef100_Q90YB5 AvEna neural variant n=1 Tax=Gallus gallus RepI... 53 9e-06
UniRef100_O93263 Avena n=1 Tax=Gallus gallus RepID=O93263_CHICK 53 9e-06
UniRef100_Q4A371 Putative membrane protein n=2 Tax=Emiliania hux... 53 9e-06
UniRef100_B9EK88 Zinc finger protein 318 n=1 Tax=Mus musculus Re... 53 9e-06
UniRef100_B0V2M3 Zinc finger protein 318 n=3 Tax=Mus musculus Re... 53 9e-06
UniRef100_Q8YQB7 All3916 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 53 9e-06
UniRef100_C2B0M4 Putative uncharacterized protein n=1 Tax=Citrob... 53 9e-06
UniRef100_Q013M1 Chromosome 08 contig 1, DNA sequence n=1 Tax=Os... 53 9e-06
UniRef100_C5YPR8 Putative uncharacterized protein Sb08g017900 n=... 53 9e-06
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 53 9e-06