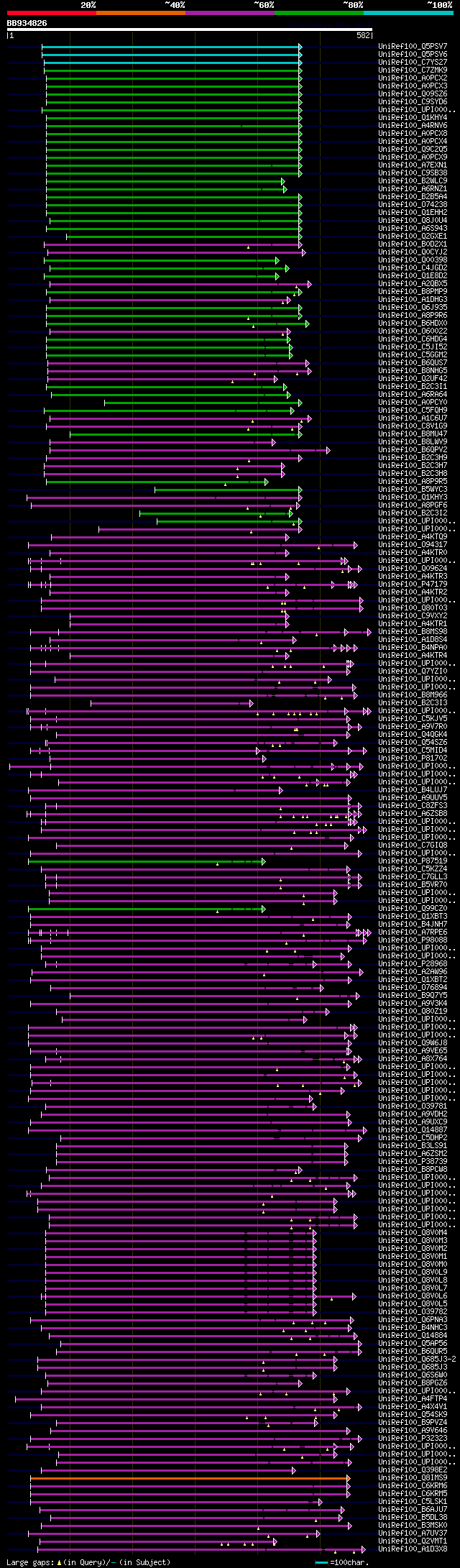
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB934826 RCC07528
(582 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q5PSV7 Snodprot-FG n=1 Tax=Gibberella zeae RepID=Q5PSV... 264 4e-69
UniRef100_Q5PSV6 Snodprot-FS n=1 Tax=Gibberella pulicaris RepID=... 256 9e-67
UniRef100_C7YS27 Putative uncharacterized protein n=1 Tax=Nectri... 204 4e-51
UniRef100_C7ZMK9 Putative uncharacterized protein n=1 Tax=Nectri... 180 8e-44
UniRef100_A0PCX2 Epl1 protein n=1 Tax=Trichoderma atroviride Rep... 179 1e-43
UniRef100_A0PCX3 Epl1 protein n=1 Tax=Trichoderma asperellum Rep... 178 3e-43
UniRef100_Q09SZ6 Eliciting plant response-like protein n=1 Tax=H... 176 9e-43
UniRef100_C9SYD6 SnodProt1 n=1 Tax=Verticillium albo-atrum VaMs.... 176 9e-43
UniRef100_UPI000023EB97 hypothetical protein FG11205.1 n=1 Tax=G... 175 3e-42
UniRef100_Q1KHY4 Snodprot1 n=1 Tax=Hypocrea virens RepID=Q1KHY4_... 174 3e-42
UniRef100_A4RNV6 Putative uncharacterized protein n=1 Tax=Magnap... 174 6e-42
UniRef100_A0PCX8 Epl1 protein n=1 Tax=Trichoderma viride RepID=A... 170 6e-41
UniRef100_A0PCX4 Epl1 protein n=3 Tax=Hypocreaceae RepID=A0PCX4_... 169 1e-40
UniRef100_Q9C2Q5 Probable SnodProt1 n=1 Tax=Neurospora crassa Re... 162 2e-38
UniRef100_A0PCX9 Epl1 protein n=1 Tax=Trichoderma viride RepID=A... 161 4e-38
UniRef100_A7EXN1 Putative uncharacterized protein n=1 Tax=Sclero... 158 3e-37
UniRef100_C9SB38 SnodProt1 n=1 Tax=Verticillium albo-atrum VaMs.... 157 4e-37
UniRef100_B2WLC9 Heat-stable 19 kDa antigen n=1 Tax=Pyrenophora ... 151 3e-35
UniRef100_A6RNZ1 Putative uncharacterized protein n=1 Tax=Botryo... 151 3e-35
UniRef100_B2B5A4 Predicted CDS Pa_2_4030 n=1 Tax=Podospora anser... 149 2e-34
UniRef100_O74238 Protein SnodProt1 n=1 Tax=Phaeosphaeria nodorum... 149 2e-34
UniRef100_Q1EHH2 Asp f 13-like protein n=1 Tax=Cochliobolus luna... 146 1e-33
UniRef100_Q8J0U4 Secreted protein 1 n=1 Tax=Leptosphaeria macula... 138 4e-31
UniRef100_A6S943 Putative uncharacterized protein n=1 Tax=Botryo... 137 8e-31
UniRef100_Q2GXE1 Putative uncharacterized protein n=1 Tax=Chaeto... 132 1e-29
UniRef100_B0D2X1 Cerato-platanin-related secreted protein n=1 Ta... 129 2e-28
UniRef100_Q0CYJ2 Allergen Asp f 15 n=1 Tax=Aspergillus terreus N... 128 4e-28
UniRef100_Q00398 Heat-stable 19 kDa antigen n=1 Tax=Coccidioides... 127 6e-28
UniRef100_C4JGD2 Heat-stable 19 kDa antigen n=1 Tax=Uncinocarpus... 126 1e-27
UniRef100_Q1E8D2 Heat-stable 19 kDa antigen n=1 Tax=Coccidioides... 126 1e-27
UniRef100_A2QBX5 Function: CS-Ag from C. immitis is a heat-stabl... 125 2e-27
UniRef100_B8PMP9 Predicted protein n=1 Tax=Postia placenta Mad-6... 124 4e-27
UniRef100_A1DHG3 Allergen Asp F13 n=1 Tax=Neosartorya fischeri N... 122 3e-26
UniRef100_Q6J935 Immunomodulatory protein n=1 Tax=Taiwanofungus ... 121 3e-26
UniRef100_A8P9R6 Putative uncharacterized protein n=1 Tax=Coprin... 120 8e-26
UniRef100_B6HDX0 Pc20g15140 protein n=1 Tax=Penicillium chrysoge... 120 1e-25
UniRef100_O60022 Allergen Asp f 15 n=2 Tax=Aspergillus fumigatus... 119 1e-25
UniRef100_C6HDG4 CS antigen n=2 Tax=Ajellomyces capsulatus RepID... 118 4e-25
UniRef100_C5JI52 Allergenic cerato-platanin Asp F13 n=1 Tax=Ajel... 117 6e-25
UniRef100_C5GGM2 Allergenic cerato-platanin Asp F13 n=1 Tax=Ajel... 117 6e-25
UniRef100_B6QUS7 Heat-stable 19 kDa antigen, putative n=1 Tax=Pe... 115 2e-24
UniRef100_B8NHG5 Allergenic cerato-platanin Asp F13 n=1 Tax=Aspe... 113 9e-24
UniRef100_Q2UF42 Predicted protein n=1 Tax=Aspergillus oryzae Re... 113 1e-23
UniRef100_B2C3I1 Cerato-platanin-like protein n=1 Tax=Moniliopht... 113 1e-23
UniRef100_A6RA64 Predicted protein n=1 Tax=Ajellomyces capsulatu... 111 5e-23
UniRef100_A0PCY0 Epl2 protein (Fragment) n=1 Tax=Trichoderma atr... 110 6e-23
UniRef100_C5FQH9 Putative uncharacterized protein n=1 Tax=Micros... 110 1e-22
UniRef100_A1C6U7 Allergen Asp F13 n=1 Tax=Aspergillus clavatus R... 107 9e-22
UniRef100_C8V1G9 Allergenic cerato-platanin Asp F13 (AFU_ortholo... 105 2e-21
UniRef100_B8MU47 Allergenic cerato-platanin Asp F13 n=1 Tax=Tala... 105 2e-21
UniRef100_B8LWV9 Heat-stable 19 kDa antigen, putative n=1 Tax=Ta... 105 2e-21
UniRef100_B6QPV2 Allergenic cerato-platanin Asp F13 n=1 Tax=Peni... 104 4e-21
UniRef100_B2C3H9 Cerato-platanin-like protein n=2 Tax=Moniliopht... 104 4e-21
UniRef100_B2C3H7 Cerato-platanin 1 n=1 Tax=Moniliophthora pernic... 99 2e-19
UniRef100_B2C3H8 Cerato-platanin 1 n=2 Tax=Moniliophthora pernic... 99 2e-19
UniRef100_A8P9R5 Putative uncharacterized protein n=1 Tax=Coprin... 98 4e-19
UniRef100_B5WYC3 Putative uncharacterized protein (Fragment) n=1... 97 9e-19
UniRef100_Q1KHY3 Snodprot2 n=1 Tax=Hypocrea virens RepID=Q1KHY3_... 89 3e-16
UniRef100_A8PGF6 Putative uncharacterized protein n=1 Tax=Coprin... 83 2e-14
UniRef100_B2C3I2 Cerato-platanin-like protein (Fragment) n=1 Tax... 76 2e-12
UniRef100_UPI000187EC69 hypothetical protein MPER_12672 n=1 Tax=... 73 1e-11
UniRef100_UPI000187C8B5 hypothetical protein MPER_03016 n=1 Tax=... 71 7e-11
UniRef100_A4KTQ9 Cerato-populin n=1 Tax=Ceratocystis populicola ... 70 2e-10
UniRef100_O94317 Uncharacterized serine-rich protein C215.13 n=1... 67 8e-10
UniRef100_A4KTR0 CF-CRO protein n=1 Tax=Ceratocystis fimbriata R... 66 2e-09
UniRef100_UPI00017C2D67 PREDICTED: hypothetical protein isoform ... 65 3e-09
UniRef100_Q09624 Location of vulva defective 1 n=1 Tax=Caenorhab... 65 4e-09
UniRef100_A4KTR3 CF-IPO protein n=1 Tax=Ceratocystis fimbriata R... 65 5e-09
UniRef100_P47179 Cell wall protein DAN4 n=1 Tax=Saccharomyces ce... 65 5e-09
UniRef100_A4KTR2 CF-COF protein n=1 Tax=Ceratocystis fimbriata R... 64 9e-09
UniRef100_UPI00015DF142 mucin 6, gastric n=1 Tax=Mus musculus Re... 64 9e-09
UniRef100_Q80T03 Mucin-6 n=1 Tax=Mus musculus RepID=MUC6_MOUSE 64 9e-09
UniRef100_C9VXY2 Cerato-variosporin n=1 Tax=Ceratocystis variosp... 64 1e-08
UniRef100_A4KTR1 CF-FAG protein n=1 Tax=Ceratocystis fimbriata R... 63 2e-08
UniRef100_B8MS98 Putative uncharacterized protein n=1 Tax=Talaro... 62 3e-08
UniRef100_A1D8S4 Putative uncharacterized protein n=1 Tax=Neosar... 62 4e-08
UniRef100_B4NPA0 GK17577 n=1 Tax=Drosophila willistoni RepID=B4N... 61 6e-08
UniRef100_A4KTR4 CF-THEO protein n=1 Tax=Ceratocystis cacaofunes... 60 9e-08
UniRef100_UPI000175F22A PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_Q7YZI0 MBCTL1 (Fragment) n=1 Tax=Monosiga brevicollis ... 60 1e-07
UniRef100_UPI0001A2BBAD Mucin-2 precursor (Intestinal mucin-2). ... 60 1e-07
UniRef100_UPI0001A2B8C7 UPI0001A2B8C7 related cluster n=1 Tax=Da... 60 1e-07
UniRef100_B8M966 Putative uncharacterized protein n=1 Tax=Talaro... 60 1e-07
UniRef100_B2C3I3 Cerato-platanin-like protein (Fragment) n=1 Tax... 60 2e-07
UniRef100_UPI0001553895 PREDICTED: similar to C6orf205 protein n... 59 2e-07
UniRef100_C5KJV5 Glycoprotein X, putative n=1 Tax=Perkinsus mari... 59 2e-07
UniRef100_A9V7R0 Predicted protein n=1 Tax=Monosiga brevicollis ... 59 3e-07
UniRef100_Q4QGK4 Surface antigen protein 2, putative n=1 Tax=Lei... 59 4e-07
UniRef100_Q54SZ6 Putative uncharacterized protein DDB_G0282133 n... 59 4e-07
UniRef100_C5MID4 Predicted protein n=1 Tax=Candida tropicalis MY... 58 5e-07
UniRef100_P81702 Cerato-platanin n=2 Tax=Ceratocystis platani Re... 58 6e-07
UniRef100_UPI00017C3B3D PREDICTED: similar to mucin 2, partial n... 58 6e-07
UniRef100_UPI0001761229 PREDICTED: hypothetical protein, partial... 58 6e-07
UniRef100_UPI0001A2DE84 UPI0001A2DE84 related cluster n=1 Tax=Da... 58 6e-07
UniRef100_B4LUJ7 GJ23908 n=1 Tax=Drosophila virilis RepID=B4LUJ7... 58 6e-07
UniRef100_A9UUV5 Predicted protein n=1 Tax=Monosiga brevicollis ... 58 6e-07
UniRef100_C8ZFS3 Aga1p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 58 6e-07
UniRef100_A6ZSB8 A-agglutinin anchorage subunit n=1 Tax=Saccharo... 58 6e-07
UniRef100_UPI00019246D2 PREDICTED: hypothetical protein n=1 Tax=... 57 8e-07
UniRef100_UPI00015537E3 PREDICTED: similar to C6orf205 protein n... 57 8e-07
UniRef100_UPI0000DB789A PREDICTED: similar to CG12807-PA n=1 Tax... 57 8e-07
UniRef100_C7GIQ8 Wsc4p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 57 8e-07
UniRef100_UPI000023EB6E hypothetical protein FG10435.1 n=1 Tax=G... 57 1e-06
UniRef100_P87519 GP80 n=1 Tax=Bovine herpesvirus 4 RepID=P87519_... 57 1e-06
UniRef100_C5KZZ4 Putative uncharacterized protein n=1 Tax=Perkin... 57 1e-06
UniRef100_C7GLL3 Aga1p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 57 1e-06
UniRef100_B5VR70 YNR044Wp-like protein (Fragment) n=1 Tax=Saccha... 57 1e-06
UniRef100_UPI00017B3035 UPI00017B3035 related cluster n=1 Tax=Te... 57 1e-06
UniRef100_UPI00017B3034 UPI00017B3034 related cluster n=1 Tax=Te... 57 1e-06
UniRef100_Q99CZ0 GP80-like protein n=1 Tax=Bovine herpesvirus 4 ... 57 1e-06
UniRef100_Q1XBT3 GP80-like protein n=1 Tax=Bovine herpesvirus 4 ... 57 1e-06
UniRef100_B4JNH7 GH24138 n=1 Tax=Drosophila grimshawi RepID=B4JN... 57 1e-06
UniRef100_A7RPE6 Predicted protein n=1 Tax=Nematostella vectensi... 57 1e-06
UniRef100_P98088 Mucin-5AC (Fragments) n=1 Tax=Homo sapiens RepI... 57 1e-06
UniRef100_UPI000175F172 PREDICTED: similar to glycoprotein (pred... 56 2e-06
UniRef100_UPI000179F724 UPI000179F724 related cluster n=1 Tax=Bo... 56 2e-06
UniRef100_P28968 Glycoprotein gp2 n=1 Tax=Equine herpesvirus typ... 56 2e-06
UniRef100_A2AW96 Novel protein containing SEA domains n=1 Tax=Da... 56 2e-06
UniRef100_Q1XBT2 GP80-like protein n=1 Tax=Bovine herpesvirus 4 ... 56 2e-06
UniRef100_O76894 CG14796 n=1 Tax=Drosophila melanogaster RepID=O... 56 2e-06
UniRef100_B9Q7Y5 Poly(A) polymerase, putative n=1 Tax=Toxoplasma... 56 2e-06
UniRef100_A9V3K4 Predicted protein n=1 Tax=Monosiga brevicollis ... 56 2e-06
UniRef100_Q80Z19 Mucin-2 (Fragments) n=1 Tax=Mus musculus RepID=... 56 2e-06
UniRef100_UPI0001761210 PREDICTED: similar to C25F9.2 n=1 Tax=Da... 55 3e-06
UniRef100_UPI0001760FAF PREDICTED: similar to rCG47301 n=1 Tax=D... 55 3e-06
UniRef100_UPI0000E46717 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_Q9W6J8 Chimeric AFGP/trypsinogen-like serine protease ... 55 3e-06
UniRef100_A9VE65 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 3e-06
UniRef100_A8X764 Putative uncharacterized protein n=1 Tax=Caenor... 55 3e-06
UniRef100_UPI000194C51E PREDICTED: mucin 2, oligomeric mucus/gel... 55 4e-06
UniRef100_UPI0001927106 PREDICTED: hypothetical protein, partial... 55 4e-06
UniRef100_UPI0001924D7A PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0000EAFF6E C-type LECtin family member (clec-200) n... 55 4e-06
UniRef100_UPI0000E48EFF PREDICTED: hypothetical protein, partial... 55 4e-06
UniRef100_O39781 Membrane glycoprotein n=1 Tax=Equid herpesvirus... 55 4e-06
UniRef100_A9VDH2 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 4e-06
UniRef100_A9UXC9 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 4e-06
UniRef100_Q14887 Mucin (Fragment) n=1 Tax=Homo sapiens RepID=Q14... 55 4e-06
UniRef100_C5DHP2 KLTH0E05940p n=1 Tax=Lachancea thermotolerans C... 55 4e-06
UniRef100_B3LS91 Cell wall integrity and stress response compone... 55 4e-06
UniRef100_A6ZSM2 Cell wall integrity and stress response compone... 55 4e-06
UniRef100_P38739 Cell wall integrity and stress response compone... 55 4e-06
UniRef100_B8PCW8 Predicted protein n=1 Tax=Postia placenta Mad-6... 55 5e-06
UniRef100_UPI0001A5ECD4 PREDICTED: similar to mucin 2 n=1 Tax=Ho... 55 5e-06
UniRef100_UPI000176068E PREDICTED: si:dkeyp-27b10.2 n=1 Tax=Dani... 55 5e-06
UniRef100_UPI0000DA1FBD PREDICTED: hypothetical protein n=1 Tax=... 55 5e-06
UniRef100_UPI0001AE79CA UPI0001AE79CA related cluster n=1 Tax=Ho... 55 5e-06
UniRef100_UPI0000D626C2 mucin 17 n=1 Tax=Homo sapiens RepID=UPI0... 55 5e-06
UniRef100_UPI00006C10F2 UPI00006C10F2 related cluster n=1 Tax=Ho... 55 5e-06
UniRef100_UPI00001B03AD Mucin-2 precursor (Intestinal mucin-2). ... 55 5e-06
UniRef100_Q8V0M4 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0M3 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0M2 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0M1 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0M0 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0L9 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0L8 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0L7 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0L6 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 5e-06
UniRef100_Q8V0L5 Glycoprotein gp2 n=1 Tax=Equid herpesvirus 1 Re... 55 5e-06
UniRef100_O39782 Membrane glycoprotein n=1 Tax=Equid herpesvirus... 55 5e-06
UniRef100_Q6PNA3 Omega gliadin n=1 Tax=Triticum aestivum RepID=Q... 55 5e-06
UniRef100_B4NHC3 GK13047 n=1 Tax=Drosophila willistoni RepID=B4N... 55 5e-06
UniRef100_Q14884 Intestinal mucin (Fragment) n=1 Tax=Homo sapien... 55 5e-06
UniRef100_Q5AP56 Putative uncharacterized protein n=1 Tax=Candid... 55 5e-06
UniRef100_B6QUR5 Putative uncharacterized protein n=1 Tax=Penici... 55 5e-06
UniRef100_Q685J3-2 Isoform 2 of Mucin-17 n=1 Tax=Homo sapiens Re... 55 5e-06
UniRef100_Q685J3 Mucin-17 n=1 Tax=Homo sapiens RepID=MUC17_HUMAN 55 5e-06
UniRef100_Q6S6W0 Glycoprotein gp2 n=1 Tax=Equine herpesvirus 1 (... 55 5e-06
UniRef100_B8PGZ6 Predicted protein n=1 Tax=Postia placenta Mad-6... 54 7e-06
UniRef100_UPI0001A2B8DA UPI0001A2B8DA related cluster n=1 Tax=Da... 54 7e-06
UniRef100_A4FTP4 Putative uncharacterized protein n=1 Tax=Cyprin... 54 7e-06
UniRef100_A4X4V1 Putative uncharacterized protein n=1 Tax=Salini... 54 7e-06
UniRef100_Q54SK9 Putative uncharacterized protein n=1 Tax=Dictyo... 54 7e-06
UniRef100_B9PVZ4 Putative uncharacterized protein n=1 Tax=Toxopl... 54 7e-06
UniRef100_A9V646 Predicted protein n=1 Tax=Monosiga brevicollis ... 54 7e-06
UniRef100_P32323 A-agglutinin anchorage subunit n=1 Tax=Saccharo... 54 7e-06
UniRef100_UPI0000E59252 mucin 2 precursor n=1 Tax=Homo sapiens R... 54 9e-06
UniRef100_UPI0000E47593 PREDICTED: similar to LOC594926 protein ... 54 9e-06
UniRef100_UPI0000079EF9 hypothetical protein Y51B11A.1 n=1 Tax=C... 54 9e-06
UniRef100_Q398E2 Membrane protein-like n=1 Tax=Burkholderia sp. ... 54 9e-06
UniRef100_Q8IMS9 CG31439 n=1 Tax=Drosophila melanogaster RepID=Q... 54 9e-06
UniRef100_C6KRM6 Protein C30H6.1b, partially confirmed by transc... 54 9e-06
UniRef100_C6KRM5 Protein C30H6.1a, partially confirmed by transc... 54 9e-06
UniRef100_C5LSK1 Cell wall integrity and stress response compone... 54 9e-06
UniRef100_B6AJU7 Putative uncharacterized protein n=1 Tax=Crypto... 54 9e-06
UniRef100_B5DL38 GA22995 n=1 Tax=Drosophila pseudoobscura pseudo... 54 9e-06
UniRef100_B3MSK0 GF21485 n=1 Tax=Drosophila ananassae RepID=B3MS... 54 9e-06
UniRef100_A7UV37 AGAP009669-PA (Fragment) n=1 Tax=Anopheles gamb... 54 9e-06
UniRef100_Q2VMT1 Phenylalanine ammonia lyase (Fragment) n=1 Tax=... 54 9e-06
UniRef100_A1D3X8 Putative uncharacterized protein n=1 Tax=Neosar... 54 9e-06
UniRef100_Q02817 Mucin-2 n=1 Tax=Homo sapiens RepID=MUC2_HUMAN 54 9e-06