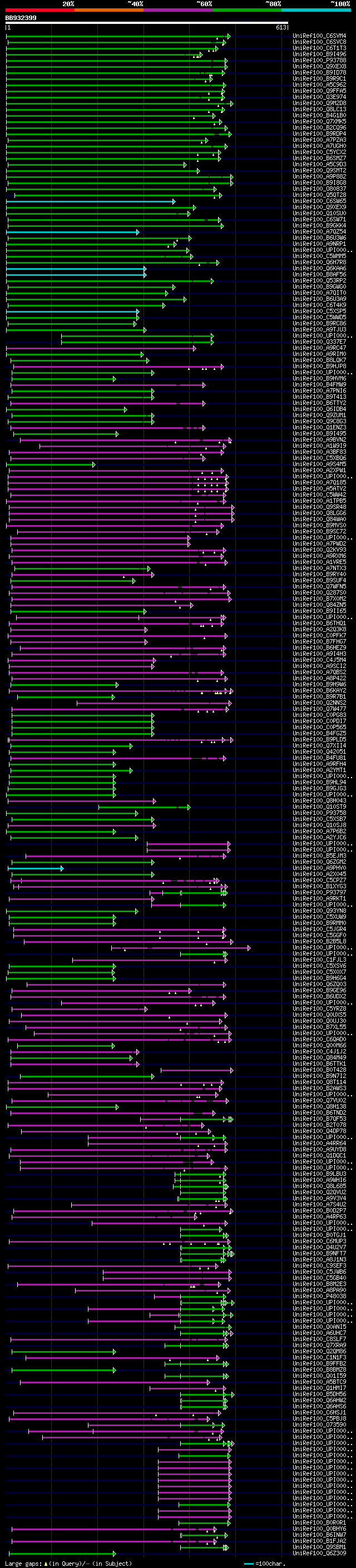
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB932399 RCC04335
(613 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_C6SVM4 Putative uncharacterized protein n=1 Tax=Glycin... 208 2e-52
UniRef100_C6SVC8 Putative uncharacterized protein n=1 Tax=Glycin... 207 7e-52
UniRef100_C6T1T3 Putative uncharacterized protein n=1 Tax=Glycin... 206 9e-52
UniRef100_B9I496 Predicted protein n=1 Tax=Populus trichocarpa R... 201 4e-50
UniRef100_P93788 Remorin n=1 Tax=Solanum tuberosum RepID=REMO_SOLTU 199 1e-49
UniRef100_Q9XEX8 Remorin 1 n=1 Tax=Solanum lycopersicum RepID=Q9... 199 2e-49
UniRef100_B9ID78 Predicted protein n=1 Tax=Populus trichocarpa R... 194 5e-48
UniRef100_B9R9C1 Remorin, putative n=1 Tax=Ricinus communis RepI... 193 1e-47
UniRef100_A5C962 Putative uncharacterized protein n=1 Tax=Vitis ... 192 2e-47
UniRef100_Q9FFA5 Similarity to remorin n=1 Tax=Arabidopsis thali... 191 4e-47
UniRef100_Q3E974 Putative uncharacterized protein At5g23750.2 n=... 190 7e-47
UniRef100_Q9M2D8 Uncharacterized protein At3g61260 n=1 Tax=Arabi... 190 9e-47
UniRef100_Q8LC13 Remorin n=1 Tax=Arabidopsis thaliana RepID=Q8LC... 189 2e-46
UniRef100_B4G1B0 Putative uncharacterized protein n=1 Tax=Zea ma... 184 5e-45
UniRef100_Q7XMK5 Os04g0533300 protein n=2 Tax=Oryza sativa RepID... 182 2e-44
UniRef100_B2CQ96 Putative remorin a3b4 n=1 Tax=Solanum tuberosum... 181 3e-44
UniRef100_B9RDP4 Remorin, putative n=1 Tax=Ricinus communis RepI... 181 6e-44
UniRef100_A7PZA3 Chromosome chr15 scaffold_40, whole genome shot... 175 3e-42
UniRef100_A7UGH0 Putative remorin a4-e8 n=1 Tax=Solanum tuberosu... 174 4e-42
UniRef100_C5YCX2 Putative uncharacterized protein Sb06g023630 n=... 173 9e-42
UniRef100_B6SMZ7 Remorin n=1 Tax=Zea mays RepID=B6SMZ7_MAIZE 173 1e-41
UniRef100_A5C9D3 Putative uncharacterized protein n=1 Tax=Vitis ... 172 3e-41
UniRef100_Q9SMT2 Remorin-like protein n=1 Tax=Arabidopsis thalia... 171 3e-41
UniRef100_A9P882 Predicted protein n=1 Tax=Populus trichocarpa R... 171 3e-41
UniRef100_B9I8G8 Predicted protein n=1 Tax=Populus trichocarpa R... 170 1e-40
UniRef100_O80837 Remorin n=1 Tax=Arabidopsis thaliana RepID=REMO... 168 4e-40
UniRef100_Q5QT28 Putative remorin 1 protein (Fragment) n=1 Tax=O... 164 7e-39
UniRef100_C6SW65 Putative uncharacterized protein n=1 Tax=Glycin... 160 6e-38
UniRef100_Q9XEX9 Remorin 2 n=1 Tax=Solanum lycopersicum RepID=Q9... 155 2e-36
UniRef100_Q10SU0 Os03g0111200 protein n=2 Tax=Oryza sativa RepID... 151 5e-35
UniRef100_C6SW71 Putative uncharacterized protein n=1 Tax=Glycin... 150 8e-35
UniRef100_B9GKK4 Predicted protein n=1 Tax=Populus trichocarpa R... 149 1e-34
UniRef100_A7QZ54 Chromosome undetermined scaffold_265, whole gen... 149 1e-34
UniRef100_B6U3W6 Remorin n=1 Tax=Zea mays RepID=B6U3W6_MAIZE 149 2e-34
UniRef100_A9NRP1 Putative uncharacterized protein n=1 Tax=Picea ... 148 3e-34
UniRef100_UPI0001982C2F PREDICTED: hypothetical protein n=1 Tax=... 146 2e-33
UniRef100_C5WMM5 Putative uncharacterized protein Sb01g049810 n=... 143 1e-32
UniRef100_Q6H7R8 Os02g0642200 protein n=2 Tax=Oryza sativa RepID... 142 3e-32
UniRef100_Q6KAA6 Os02g0824500 protein n=1 Tax=Oryza sativa Japon... 140 1e-31
UniRef100_B8AF56 Putative uncharacterized protein n=1 Tax=Oryza ... 140 1e-31
UniRef100_Q53RP2 Putative uncharacterized protein n=2 Tax=Oryza ... 139 2e-31
UniRef100_B9GWG0 Predicted protein n=1 Tax=Populus trichocarpa R... 139 2e-31
UniRef100_A7QIT0 Chromosome chr2 scaffold_105, whole genome shot... 136 1e-30
UniRef100_B6U3A9 Remorin n=1 Tax=Zea mays RepID=B6U3A9_MAIZE 132 2e-29
UniRef100_C6T4K9 Putative uncharacterized protein n=1 Tax=Glycin... 125 3e-27
UniRef100_C5XSP5 Putative uncharacterized protein Sb04g033660 n=... 125 3e-27
UniRef100_C5WWD5 Putative uncharacterized protein Sb01g017740 n=... 115 3e-24
UniRef100_B9RC86 Remorin, putative n=1 Tax=Ricinus communis RepI... 107 6e-22
UniRef100_A9TJU3 Predicted protein n=1 Tax=Physcomitrella patens... 98 5e-19
UniRef100_UPI0000DD9A28 Os10g0503800 n=1 Tax=Oryza sativa Japoni... 97 1e-18
UniRef100_Q337E7 cDNA clone:J033075A20, full insert sequence n=1... 97 1e-18
UniRef100_A9RC47 Predicted protein n=1 Tax=Physcomitrella patens... 94 7e-18
UniRef100_A9RIM0 Predicted protein (Fragment) n=1 Tax=Physcomitr... 91 1e-16
UniRef100_B8LQK7 Putative uncharacterized protein n=1 Tax=Picea ... 81 6e-14
UniRef100_B9HJP8 Predicted protein n=1 Tax=Populus trichocarpa R... 80 1e-13
UniRef100_UPI0001982A11 PREDICTED: similar to remorin family pro... 78 7e-13
UniRef100_B9HVM6 Predicted protein n=1 Tax=Populus trichocarpa R... 78 7e-13
UniRef100_B4FMW9 Putative uncharacterized protein n=1 Tax=Zea ma... 78 7e-13
UniRef100_A7PNI6 Chromosome chr1 scaffold_22, whole genome shotg... 78 7e-13
UniRef100_B9T413 DNA binding protein, putative n=1 Tax=Ricinus c... 77 9e-13
UniRef100_B6TTY2 DNA binding protein n=1 Tax=Zea mays RepID=B6TT... 77 1e-12
UniRef100_Q6IDB4 At4g00670 n=1 Tax=Arabidopsis thaliana RepID=Q6... 76 2e-12
UniRef100_Q9ZUM1 Expressed protein n=1 Tax=Arabidopsis thaliana ... 76 2e-12
UniRef100_Q9C8G3 Putative uncharacterized protein T4K22.7 n=1 Ta... 75 4e-12
UniRef100_Q1ENZ3 Remorin-related n=1 Tax=Musa acuminata RepID=Q1... 75 6e-12
UniRef100_B9I495 Predicted protein n=1 Tax=Populus trichocarpa R... 74 7e-12
UniRef100_A9BVN2 Protein TolA n=1 Tax=Delftia acidovorans SPH-1 ... 74 1e-11
UniRef100_A1W9I9 TonB family protein n=2 Tax=Comamonadaceae RepI... 74 1e-11
UniRef100_A3BF83 Putative uncharacterized protein n=1 Tax=Oryza ... 74 1e-11
UniRef100_C5XBQ6 Putative uncharacterized protein Sb02g036810 n=... 73 2e-11
UniRef100_A9S4M5 Predicted protein n=1 Tax=Physcomitrella patens... 73 2e-11
UniRef100_A2XPW1 Putative uncharacterized protein n=1 Tax=Oryza ... 73 2e-11
UniRef100_UPI00019842F0 PREDICTED: hypothetical protein n=1 Tax=... 73 2e-11
UniRef100_A7Q185 Chromosome chr10 scaffold_43, whole genome shot... 73 2e-11
UniRef100_A5ATV2 Putative uncharacterized protein n=1 Tax=Vitis ... 73 2e-11
UniRef100_C5WW42 Putative uncharacterized protein Sb01g004200 n=... 72 3e-11
UniRef100_A1TPB5 TonB family protein n=1 Tax=Acidovorax citrulli... 72 4e-11
UniRef100_Q9SR48 F12A21.28 n=1 Tax=Arabidopsis thaliana RepID=Q9... 72 4e-11
UniRef100_Q8LGG6 Putative uncharacterized protein n=1 Tax=Arabid... 72 4e-11
UniRef100_Q84WA0 Putative uncharacterized protein At1g67590 (Fra... 72 4e-11
UniRef100_B9MVS0 Predicted protein (Fragment) n=1 Tax=Populus tr... 72 4e-11
UniRef100_B9SC72 DNA binding protein, putative n=1 Tax=Ricinus c... 72 5e-11
UniRef100_UPI0001983DC3 PREDICTED: hypothetical protein n=1 Tax=... 71 6e-11
UniRef100_A7PWD2 Chromosome chr8 scaffold_34, whole genome shotg... 71 6e-11
UniRef100_Q2KV93 TolA protein n=1 Tax=Bordetella avium 197N RepI... 70 1e-10
UniRef100_A9RXM6 Predicted protein n=1 Tax=Physcomitrella patens... 70 1e-10
UniRef100_A1VRE5 TonB family protein n=1 Tax=Polaromonas naphtha... 70 1e-10
UniRef100_A7NTX3 Chromosome chr18 scaffold_1, whole genome shotg... 70 1e-10
UniRef100_B9RY40 DNA binding protein, putative n=1 Tax=Ricinus c... 70 2e-10
UniRef100_B9SUF4 Remorin, putative n=1 Tax=Ricinus communis RepI... 69 2e-10
UniRef100_Q7WFN5 Proline-rich inner membrane protein n=1 Tax=Bor... 69 3e-10
UniRef100_Q287S0 ORF1629 n=1 Tax=Agrotis segetum nucleopolyhedro... 69 4e-10
UniRef100_B7X0M2 Protein TolA n=1 Tax=Comamonas testosteroni KF-... 69 4e-10
UniRef100_Q84ZN5 Os07g0208600 protein n=1 Tax=Oryza sativa Japon... 69 4e-10
UniRef100_B9II65 Predicted protein n=1 Tax=Populus trichocarpa R... 69 4e-10
UniRef100_UPI00019259C5 PREDICTED: hypothetical protein n=1 Tax=... 68 5e-10
UniRef100_B6THQ1 DNA binding protein n=1 Tax=Zea mays RepID=B6TH... 68 5e-10
UniRef100_A2Q3K8 Remorin, C-terminal region n=1 Tax=Medicago tru... 68 5e-10
UniRef100_C0PFK7 Putative uncharacterized protein n=1 Tax=Zea ma... 68 7e-10
UniRef100_B7FHG7 Putative uncharacterized protein n=1 Tax=Medica... 68 7e-10
UniRef100_B6HEZ9 Pc20g08960 protein n=1 Tax=Penicillium chrysoge... 68 7e-10
UniRef100_A9I4H3 Colicin exporter n=1 Tax=Bordetella petrii DSM ... 67 9e-10
UniRef100_C4J5M4 Putative uncharacterized protein n=1 Tax=Zea ma... 67 9e-10
UniRef100_A9SCI2 Predicted protein n=1 Tax=Physcomitrella patens... 67 9e-10
UniRef100_A7QBS2 Chromosome chr1 scaffold_75, whole genome shotg... 67 9e-10
UniRef100_A8P422 Predicted protein n=1 Tax=Coprinopsis cinerea o... 67 9e-10
UniRef100_B9H9W6 Predicted protein (Fragment) n=1 Tax=Populus tr... 67 2e-09
UniRef100_B6KAY2 Putative uncharacterized protein n=2 Tax=Toxopl... 67 2e-09
UniRef100_B9R7B1 DNA binding protein, putative n=1 Tax=Ricinus c... 66 2e-09
UniRef100_Q2NNS2 1629capsid n=1 Tax=Hyphantria cunea nucleopolyh... 66 3e-09
UniRef100_Q7W477 Proline-rich inner membrane protein n=1 Tax=Bor... 66 3e-09
UniRef100_C0PG83 Putative uncharacterized protein n=1 Tax=Zea ma... 66 3e-09
UniRef100_C0PDI7 Putative uncharacterized protein n=1 Tax=Zea ma... 66 3e-09
UniRef100_C0P565 Putative uncharacterized protein n=1 Tax=Zea ma... 66 3e-09
UniRef100_B4FGZ5 Putative uncharacterized protein n=1 Tax=Zea ma... 66 3e-09
UniRef100_B9PLD5 Putative uncharacterized protein n=1 Tax=Toxopl... 66 3e-09
UniRef100_Q7XII4 Os07g0569100 protein n=1 Tax=Oryza sativa Japon... 65 3e-09
UniRef100_Q42051 DNA-binding protein (Fragment) n=1 Tax=Arabidop... 65 3e-09
UniRef100_B4FU81 Putative uncharacterized protein n=1 Tax=Zea ma... 65 3e-09
UniRef100_A9RFH4 Predicted protein n=1 Tax=Physcomitrella patens... 65 3e-09
UniRef100_A2YMT1 Putative uncharacterized protein n=1 Tax=Oryza ... 65 3e-09
UniRef100_UPI0001982B80 PREDICTED: hypothetical protein n=1 Tax=... 65 4e-09
UniRef100_B9HL94 Predicted protein n=1 Tax=Populus trichocarpa R... 65 4e-09
UniRef100_B9GJG3 Predicted protein (Fragment) n=1 Tax=Populus tr... 65 4e-09
UniRef100_UPI0001983ECA PREDICTED: hypothetical protein n=1 Tax=... 65 6e-09
UniRef100_Q8H043 Putative uncharacterized protein OJ1263H11.13 n... 65 6e-09
UniRef100_Q10ST9 Remorin, C-terminal region family protein, expr... 65 6e-09
UniRef100_P93758 Putative DNA binding protein n=1 Tax=Arabidopsi... 65 6e-09
UniRef100_C5XSB7 Putative uncharacterized protein Sb04g001240 n=... 65 6e-09
UniRef100_Q10SJ8 Os03g0120200 protein n=3 Tax=Oryza sativa RepID... 65 6e-09
UniRef100_A7P6B2 Chromosome chr9 scaffold_7, whole genome shotgu... 65 6e-09
UniRef100_A2YJC6 Putative uncharacterized protein n=1 Tax=Oryza ... 65 6e-09
UniRef100_UPI0000E4A382 PREDICTED: similar to DNA-dependent prot... 64 8e-09
UniRef100_UPI0000E47F32 PREDICTED: hypothetical protein n=1 Tax=... 64 8e-09
UniRef100_B5EJM3 Protein TolA n=2 Tax=Acidithiobacillus ferrooxi... 64 8e-09
UniRef100_Q6ZGM2 Os02g0116800 protein n=1 Tax=Oryza sativa Japon... 64 8e-09
UniRef100_A9PHV0 Putative uncharacterized protein n=1 Tax=Populu... 64 8e-09
UniRef100_A2X045 Putative uncharacterized protein n=1 Tax=Oryza ... 64 8e-09
UniRef100_C5CPZ7 Protein TolA n=1 Tax=Variovorax paradoxus S110 ... 64 1e-08
UniRef100_B1XYG3 Protein TolA n=1 Tax=Leptothrix cholodnii SP-6 ... 64 1e-08
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 64 1e-08
UniRef100_A9RKT1 Predicted protein n=1 Tax=Physcomitrella patens... 64 1e-08
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 64 1e-08
UniRef100_Q93YN8 Putative uncharacterized protein T8H10.14 n=2 T... 64 1e-08
UniRef100_C5XUW9 Putative uncharacterized protein Sb04g022970 n=... 64 1e-08
UniRef100_B9RMM0 Putative uncharacterized protein n=1 Tax=Ricinu... 64 1e-08
UniRef100_C5JGR4 Putative uncharacterized protein n=1 Tax=Ajello... 64 1e-08
UniRef100_C5GGF0 Putative uncharacterized protein n=1 Tax=Ajello... 64 1e-08
UniRef100_B2B5L8 Predicted CDS Pa_2_5060 n=1 Tax=Podospora anser... 64 1e-08
UniRef100_UPI0000F2C92D PREDICTED: similar to transcription fact... 63 2e-08
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 63 2e-08
UniRef100_C1FJL3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 63 2e-08
UniRef100_C5XSV6 Putative uncharacterized protein Sb04g034210 n=... 63 2e-08
UniRef100_C5X0X7 Putative uncharacterized protein Sb01g049190 n=... 63 2e-08
UniRef100_B9H6G4 Predicted protein n=1 Tax=Populus trichocarpa R... 63 2e-08
UniRef100_Q6ZQ03 Formin-binding protein 4 n=1 Tax=Mus musculus R... 63 2e-08
UniRef100_B9GE96 Putative uncharacterized protein n=1 Tax=Oryza ... 62 3e-08
UniRef100_B6UDX2 Remorin n=1 Tax=Zea mays RepID=B6UDX2_MAIZE 62 3e-08
UniRef100_UPI0000E7F800 PREDICTED: hypothetical protein n=1 Tax=... 62 4e-08
UniRef100_C5YRZ8 Putative uncharacterized protein Sb08g021450 n=... 62 4e-08
UniRef100_Q0UXS5 Putative uncharacterized protein n=1 Tax=Phaeos... 62 4e-08
UniRef100_Q0UJ30 Putative uncharacterized protein n=1 Tax=Phaeos... 62 4e-08
UniRef100_B7XL55 Putative uncharacterized protein n=1 Tax=Entero... 62 4e-08
UniRef100_UPI0000F2E28C PREDICTED: similar to high-mobility grou... 62 5e-08
UniRef100_C6QAD0 Protein TolA n=1 Tax=Hyphomicrobium denitrifica... 62 5e-08
UniRef100_Q00M66 Remorin n=1 Tax=Glycine max RepID=Q00M66_SOYBN 62 5e-08
UniRef100_C4J1J2 Putative uncharacterized protein n=1 Tax=Zea ma... 62 5e-08
UniRef100_Q84M49 Os03g0808300 protein n=2 Tax=Oryza sativa RepID... 62 5e-08
UniRef100_B6TTK1 DNA binding protein n=1 Tax=Zea mays RepID=B6TT... 62 5e-08
UniRef100_B0T428 Glycoside hydrolase family 16 n=1 Tax=Caulobact... 61 6e-08
UniRef100_B9N7I2 Predicted protein n=1 Tax=Populus trichocarpa R... 61 6e-08
UniRef100_Q8T114 Histone-like protein n=1 Tax=Physarum polycepha... 61 6e-08
UniRef100_B2AWS3 Actin cytoskeleton-regulatory complex protein P... 61 6e-08
UniRef100_UPI00006A0DC4 WAS protein homology region 2 domain con... 61 8e-08
UniRef100_Q7VU02 Proline-rich inner membrane protein n=1 Tax=Bor... 61 8e-08
UniRef100_Q8H138 Putative DNA binding protein (Fragment) n=1 Tax... 61 8e-08
UniRef100_B6TND2 DNA binding protein n=1 Tax=Zea mays RepID=B6TN... 61 8e-08
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 61 8e-08
UniRef100_B2T078 Protein TolA n=1 Tax=Burkholderia phytofirmans ... 60 1e-07
UniRef100_Q4DP78 Mucin-associated surface protein (MASP), putati... 60 1e-07
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 60 1e-07
UniRef100_A4RR64 Predicted protein n=1 Tax=Ostreococcus lucimari... 60 1e-07
UniRef100_A9UYD8 Predicted protein n=1 Tax=Monosiga brevicollis ... 60 1e-07
UniRef100_Q1DQC1 Actin cytoskeleton-regulatory complex protein P... 60 1e-07
UniRef100_UPI00019828B0 PREDICTED: hypothetical protein n=1 Tax=... 60 2e-07
UniRef100_UPI0000E1E81E PREDICTED: similar to Arginine/proline r... 60 2e-07
UniRef100_B9LBU3 Putative uncharacterized protein n=1 Tax=Chloro... 60 2e-07
UniRef100_A9WHI6 Putative uncharacterized protein n=1 Tax=Chloro... 60 2e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 60 2e-07
UniRef100_Q2QVU2 Transposon protein, putative, CACTA, En/Spm sub... 60 2e-07
UniRef100_A9V3V4 Predicted protein n=1 Tax=Monosiga brevicollis ... 60 2e-07
UniRef100_A7S4U2 Predicted protein n=1 Tax=Nematostella vectensi... 60 2e-07
UniRef100_B0D2P7 Predicted protein n=1 Tax=Laccaria bicolor S238... 60 2e-07
UniRef100_A4RP63 Putative uncharacterized protein n=1 Tax=Magnap... 60 2e-07
UniRef100_UPI0001554901 PREDICTED: similar to dipeptidyl peptida... 59 2e-07
UniRef100_UPI0000F2D09F PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 59 2e-07
UniRef100_C6MUP3 Chromosome segregation ATPase-like protein n=1 ... 59 2e-07
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 59 2e-07
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 59 2e-07
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 59 2e-07
UniRef100_C9SEF3 Putative uncharacterized protein n=1 Tax=Vertic... 59 2e-07
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 59 2e-07
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 59 2e-07
UniRef100_B8M2E3 Adenylyl cyclase-associated protein n=1 Tax=Tal... 59 2e-07
UniRef100_A8PA90 Putative uncharacterized protein n=1 Tax=Coprin... 59 2e-07
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 59 2e-07
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 59 3e-07
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 59 3e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 59 3e-07
UniRef100_UPI0000ECD10B Zinc finger homeobox protein 4 (Zinc fin... 59 3e-07
UniRef100_Q0ANI5 OmpA/MotB domain protein n=1 Tax=Maricaulis mar... 59 3e-07
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 59 3e-07
UniRef100_C8SLF7 Translation initiation factor IF-2 n=1 Tax=Meso... 59 3e-07
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 59 3e-07
UniRef100_Q2QM86 Os12g0613600 protein n=2 Tax=Oryza sativa Japon... 59 3e-07
UniRef100_C1N1F3 Predicted protein (Fragment) n=1 Tax=Micromonas... 59 3e-07
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 59 3e-07
UniRef100_B8BMZ8 Putative uncharacterized protein n=1 Tax=Oryza ... 59 3e-07
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 59 3e-07
UniRef100_A5BTC9 Putative uncharacterized protein n=1 Tax=Vitis ... 59 3e-07
UniRef100_Q1HMI7 Formin B n=2 Tax=Trypanosoma cruzi RepID=Q1HMI7... 59 3e-07
UniRef100_B5DH56 GA25354 n=1 Tax=Drosophila pseudoobscura pseudo... 59 3e-07
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 59 3e-07
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 59 3e-07
UniRef100_C6HSJ1 Predicted protein n=1 Tax=Ajellomyces capsulatu... 59 3e-07
UniRef100_C5PBJ8 WH2 motif family protein n=1 Tax=Coccidioides p... 59 3e-07
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 59 3e-07
UniRef100_UPI0001926925 PREDICTED: hypothetical protein n=1 Tax=... 59 4e-07
UniRef100_UPI000186AA1B hypothetical protein BRAFLDRAFT_110367 n... 59 4e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 59 4e-07
UniRef100_UPI00017C32AF PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI000175F21B PREDICTED: similar to Ras-associated and... 59 4e-07
UniRef100_UPI00015604DB PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A1831 PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A1830 PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A182F PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A182E PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A182D PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00005A182B PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI00015A6C38 UPI00015A6C38 related cluster n=1 Tax=Da... 59 4e-07
UniRef100_UPI00005A1832 PREDICTED: similar to YLP motif containi... 59 4e-07
UniRef100_UPI000179E4D3 PREDICTED: Bos taurus similar to YLP mot... 59 4e-07
UniRef100_B0R0R1 Novel protein similar to human Ras association ... 59 4e-07
UniRef100_Q0BHY6 TolA family protein n=1 Tax=Burkholderia ambifa... 59 4e-07
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 59 4e-07
UniRef100_B1FJA2 Protein TolA n=1 Tax=Burkholderia ambifaria IOP... 59 4e-07
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 59 4e-07
UniRef100_Q6Z309 Os02g0767000 protein n=1 Tax=Oryza sativa Japon... 59 4e-07
UniRef100_O65276 F6N23.13 protein n=1 Tax=Arabidopsis thaliana R... 59 4e-07
UniRef100_O23188 Putative uncharacterized protein AT4g36970 n=1 ... 59 4e-07
UniRef100_B9HTA7 Predicted protein n=1 Tax=Populus trichocarpa R... 59 4e-07
UniRef100_A9T3J1 Predicted protein n=1 Tax=Physcomitrella patens... 59 4e-07
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 59 4e-07
UniRef100_A7PLA3 Chromosome chr7 scaffold_20, whole genome shotg... 59 4e-07
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 59 4e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 59 4e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 59 4e-07
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 59 4e-07
UniRef100_A7RNZ0 Predicted protein n=1 Tax=Nematostella vectensi... 59 4e-07
UniRef100_B2VZ82 Predicted protein n=1 Tax=Pyrenophora tritici-r... 59 4e-07
UniRef100_B2AVH8 Predicted CDS Pa_7_2900 n=1 Tax=Podospora anser... 59 4e-07
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 59 4e-07
UniRef100_UPI000194BDE1 PREDICTED: zinc finger homeodomain 4 n=1... 58 5e-07
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 58 5e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 58 5e-07
UniRef100_UPI0001A2D80C UPI0001A2D80C related cluster n=1 Tax=Da... 58 5e-07
UniRef100_UPI0000F31DA4 Formin-binding protein 4 (Formin-binding... 58 5e-07
UniRef100_Q098M4 Putative uncharacterized protein n=1 Tax=Stigma... 58 5e-07
UniRef100_C6TI04 Putative uncharacterized protein n=1 Tax=Glycin... 58 5e-07
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 58 5e-07
UniRef100_C1MXF0 Predicted protein n=1 Tax=Micromonas pusilla CC... 58 5e-07
UniRef100_B8A2V2 Putative uncharacterized protein n=1 Tax=Zea ma... 58 5e-07
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 58 5e-07
UniRef100_A7QGU9 Chromosome chr16 scaffold_94, whole genome shot... 58 5e-07
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 58 5e-07
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 58 5e-07
UniRef100_Q1DQI2 Putative uncharacterized protein n=1 Tax=Coccid... 58 5e-07
UniRef100_C5PAP6 Putative uncharacterized protein n=1 Tax=Coccid... 58 5e-07
UniRef100_UPI00017EFD1A PREDICTED: similar to YLP motif containi... 58 7e-07
UniRef100_UPI000150640F YLP motif containing 1 n=1 Tax=Mus muscu... 58 7e-07
UniRef100_UPI0000F2BFBF PREDICTED: similar to bromodomain PHD fi... 58 7e-07
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 58 7e-07
UniRef100_UPI00015DEB7E YLP motif containing 1 n=1 Tax=Mus muscu... 58 7e-07
UniRef100_UPI0000D8B545 YLP motif containing 1 n=1 Tax=Mus muscu... 58 7e-07
UniRef100_B4EDB9 Possible TolA-related transport transmembrane p... 58 7e-07
UniRef100_A5V8M1 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 58 7e-07
UniRef100_Q2R164 Os11g0616300 protein n=1 Tax=Oryza sativa Japon... 58 7e-07
UniRef100_B9RQE5 Putative uncharacterized protein n=1 Tax=Ricinu... 58 7e-07
UniRef100_B9F3E9 Putative uncharacterized protein n=1 Tax=Oryza ... 58 7e-07
UniRef100_B8AJ44 Putative uncharacterized protein n=1 Tax=Oryza ... 58 7e-07
UniRef100_A2ZGB8 Putative uncharacterized protein n=1 Tax=Oryza ... 58 7e-07
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 58 7e-07
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 58 7e-07
UniRef100_B3NT91 GG18988 n=1 Tax=Drosophila erecta RepID=B3NT91_... 58 7e-07
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 58 7e-07
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 58 7e-07
UniRef100_C5FY77 Formin-like protein 20 n=1 Tax=Microsporum cani... 58 7e-07
UniRef100_C0P113 Putative uncharacterized protein n=1 Tax=Ajello... 58 7e-07
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 58 7e-07
UniRef100_Q9R0I7 YLP motif-containing protein 1 n=1 Tax=Mus musc... 58 7e-07
UniRef100_Q1ZXE2 PH domain-containing protein DDB_G0287875 n=1 T... 58 7e-07
UniRef100_UPI0000F202B4 PREDICTED: similar to tubby like protein... 57 9e-07
UniRef100_UPI00001CF1F0 formin binding protein 4 n=1 Tax=Rattus ... 57 9e-07
UniRef100_UPI000069DAE3 FH1/FH2 domain-containing protein 1 (For... 57 9e-07
UniRef100_UPI0001B7B45F Fnbp4 protein. n=1 Tax=Rattus norvegicus... 57 9e-07
UniRef100_Q06KR7 Viral capsid associated protein n=1 Tax=Anticar... 57 9e-07
UniRef100_Q2Y5G9 TonB-like n=1 Tax=Nitrosospira multiformis ATCC... 57 9e-07
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 57 9e-07
UniRef100_B1YTW8 Protein TolA n=1 Tax=Burkholderia ambifaria MC4... 57 9e-07
UniRef100_A5V4K4 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 57 9e-07
UniRef100_A2SK16 Putative uncharacterized protein n=1 Tax=Methyl... 57 9e-07
UniRef100_Q7P3A4 Hypothetical WASP N-WASP MENA proteins n=1 Tax=... 57 9e-07
UniRef100_Q94AQ0 Putative uncharacterized protein At4g36970 n=1 ... 57 9e-07
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 57 9e-07
UniRef100_Q01AC3 Meltrins, fertilins and related Zn-dependent me... 57 9e-07
UniRef100_C1E983 Predicted protein n=1 Tax=Micromonas sp. RCC299... 57 9e-07
UniRef100_A7PES6 Chromosome chr11 scaffold_13, whole genome shot... 57 9e-07
UniRef100_Q7YY78 Protease, possible n=2 Tax=Cryptosporidium parv... 57 9e-07
UniRef100_Q4CTN2 Dispersed gene family protein 1 (DGF-1), putati... 57 9e-07
UniRef100_C5KAA2 Putative uncharacterized protein n=1 Tax=Perkin... 57 9e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 57 9e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 57 9e-07
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 57 9e-07
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 57 9e-07
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 57 9e-07
UniRef100_Q6NX58 MAP7D1 protein (Fragment) n=1 Tax=Homo sapiens ... 57 9e-07
UniRef100_B5MDZ6 Putative uncharacterized protein MAP7D1 n=1 Tax... 57 9e-07
UniRef100_Q96VJ2 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 57 9e-07
UniRef100_Q6AHU5 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 57 9e-07
UniRef100_Q0U2K7 Putative uncharacterized protein n=1 Tax=Phaeos... 57 9e-07
UniRef100_A7E486 Putative uncharacterized protein n=1 Tax=Sclero... 57 9e-07
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 57 9e-07
UniRef100_Q3KQU3-2 Isoform 2 of MAP7 domain-containing protein 1... 57 9e-07
UniRef100_Q3KQU3-4 Isoform 4 of MAP7 domain-containing protein 1... 57 9e-07
UniRef100_Q3KQU3 MAP7 domain-containing protein 1 n=1 Tax=Homo s... 57 9e-07
UniRef100_UPI0000E81D65 PREDICTED: similar to keratin 7, partial... 57 1e-06
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 57 1e-06
UniRef100_UPI0001553749 PREDICTED: hypothetical protein n=1 Tax=... 57 1e-06
UniRef100_UPI0000DA3EC9 PREDICTED: similar to tumor endothelial ... 57 1e-06
UniRef100_UPI0000D99870 PREDICTED: similar to proline arginine r... 57 1e-06
UniRef100_UPI0000D9986E PREDICTED: similar to proline arginine r... 57 1e-06
UniRef100_UPI00006D5CAD PREDICTED: similar to proline arginine r... 57 1e-06
UniRef100_UPI0001B7AE60 UPI0001B7AE60 related cluster n=1 Tax=Ra... 57 1e-06
UniRef100_UPI0001B7AE51 UPI0001B7AE51 related cluster n=1 Tax=Ra... 57 1e-06
UniRef100_UPI00016EA76C UPI00016EA76C related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016EA76B UPI00016EA76B related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016EA501 UPI00016EA501 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 57 1e-06
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 57 1e-06
UniRef100_B6VC31 VP78 n=1 Tax=Iragoides fasciata nucleopolyhedro... 57 1e-06
UniRef100_Q2RTE8 Putative uncharacterized protein n=1 Tax=Rhodos... 57 1e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 57 1e-06
UniRef100_C1MRV3 Predicted protein n=1 Tax=Micromonas pusilla CC... 57 1e-06
UniRef100_C1MN00 Predicted protein n=1 Tax=Micromonas pusilla CC... 57 1e-06
UniRef100_A9RE38 Predicted protein n=1 Tax=Physcomitrella patens... 57 1e-06
UniRef100_A5AJX1 Putative uncharacterized protein n=1 Tax=Vitis ... 57 1e-06
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 57 1e-06
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 57 1e-06
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 57 1e-06
UniRef100_C1GQP5 Predicted protein n=1 Tax=Paracoccidioides bras... 57 1e-06
UniRef100_P0CB49 YLP motif-containing protein 1 n=1 Tax=Rattus n... 57 1e-06
UniRef100_UPI000176023F PREDICTED: similar to protein kinase bet... 57 2e-06
UniRef100_UPI0000D9AE8D PREDICTED: similar to microtubule-associ... 57 2e-06
UniRef100_UPI0000D9AE8C PREDICTED: similar to microtubule-associ... 57 2e-06
UniRef100_UPI0000D9AE8B PREDICTED: similar to microtubule-associ... 57 2e-06
UniRef100_UPI0000DC1294 UPI0000DC1294 related cluster n=1 Tax=Ra... 57 2e-06
UniRef100_UPI00016E3AF1 UPI00016E3AF1 related cluster n=1 Tax=Ta... 57 2e-06
UniRef100_UPI00016E3ADC UPI00016E3ADC related cluster n=1 Tax=Ta... 57 2e-06
UniRef100_UPI00016E3ADB UPI00016E3ADB related cluster n=1 Tax=Ta... 57 2e-06
UniRef100_UPI00016E3ABE UPI00016E3ABE related cluster n=1 Tax=Ta... 57 2e-06
UniRef100_Q8QTE3 WSSV198 n=1 Tax=Shrimp white spot syndrome viru... 57 2e-06
UniRef100_Q39J77 TonB/TolA-like protein n=1 Tax=Burkholderia sp.... 57 2e-06
UniRef100_C3PP34 Actin polymerization protein RickA n=1 Tax=Rick... 57 2e-06
UniRef100_A8GSX3 Putative uncharacterized protein n=1 Tax=Ricket... 57 2e-06
UniRef100_C7HY75 Protein TolA n=1 Tax=Thiomonas intermedia K12 R... 57 2e-06
UniRef100_Q5VR02 Hydroxyproline-rich glycoprotein-like n=1 Tax=O... 57 2e-06
UniRef100_Q53N23 Transposon protein, putative, CACTA, En/Spm sub... 57 2e-06
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 57 2e-06
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 57 2e-06
UniRef100_Q4D9L5 Putative uncharacterized protein n=1 Tax=Trypan... 57 2e-06
UniRef100_Q16PS7 Cxyorf1 (Fragment) n=2 Tax=Aedes aegypti RepID=... 57 2e-06
UniRef100_B4NYZ4 GE18300 n=1 Tax=Drosophila yakuba RepID=B4NYZ4_... 57 2e-06
UniRef100_A7RSP9 Predicted protein n=1 Tax=Nematostella vectensi... 57 2e-06
UniRef100_B2AXN7 Predicted CDS Pa_7_11170 n=1 Tax=Podospora anse... 57 2e-06
UniRef100_Q3KQU3-3 Isoform 3 of MAP7 domain-containing protein 1... 57 2e-06
UniRef100_A3AB67 Formin-like protein 16 n=1 Tax=Oryza sativa Jap... 57 2e-06
UniRef100_UPI00017C2CFA PREDICTED: similar to arginine/proline r... 56 2e-06
UniRef100_UPI0000E20244 PREDICTED: huntingtin n=1 Tax=Pan troglo... 56 2e-06
UniRef100_UPI0000DA3E0C PREDICTED: similar to tumor endothelial ... 56 2e-06
UniRef100_UPI000184A125 Formin-binding protein 4 (Formin-binding... 56 2e-06
UniRef100_UPI00006A0DA0 zinc finger homeodomain 4 n=1 Tax=Xenopu... 56 2e-06
UniRef100_UPI000069F8C9 RPRC1 protein n=1 Tax=Xenopus (Silurana)... 56 2e-06
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 56 2e-06
UniRef100_UPI000024DCC5 zinc finger homeodomain 4 n=1 Tax=Mus mu... 56 2e-06
UniRef100_UPI0000EE3DB8 zinc finger homeodomain 4 n=1 Tax=Homo s... 56 2e-06
UniRef100_UPI00004576BB Zinc finger homeobox protein 4 (Zinc fin... 56 2e-06
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 56 2e-06
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 56 2e-06
UniRef100_Q9WX60 Ccp protein n=1 Tax=Gluconacetobacter xylinus R... 56 2e-06
UniRef100_C6MHJ8 Protein TolA n=1 Tax=Nitrosomonas sp. AL212 Rep... 56 2e-06
UniRef100_B1TFI1 Protein TolA n=1 Tax=Burkholderia ambifaria MEX... 56 2e-06
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 56 2e-06
UniRef100_C5Y6F6 Putative uncharacterized protein Sb05g024580 n=... 56 2e-06
UniRef100_B6SIP6 DNA binding protein n=1 Tax=Zea mays RepID=B6SI... 56 2e-06
UniRef100_A7Z068 LMOD2 protein n=1 Tax=Bos taurus RepID=A7Z068_B... 56 2e-06
UniRef100_Q4CKF6 Dispersed gene family protein 1 (DGF-1), putati... 56 2e-06
UniRef100_B4G6U6 GL19111 n=1 Tax=Drosophila persimilis RepID=B4G... 56 2e-06
UniRef100_B0W487 Putative uncharacterized protein n=1 Tax=Culex ... 56 2e-06
UniRef100_C9SG64 Putative uncharacterized protein n=1 Tax=Vertic... 56 2e-06
UniRef100_A8NWX4 Putative uncharacterized protein n=1 Tax=Coprin... 56 2e-06
UniRef100_Q86UP3 Zinc finger homeobox protein 4 n=1 Tax=Homo sap... 56 2e-06
UniRef100_O94532 Formin-3 n=1 Tax=Schizosaccharomyces pombe RepI... 56 2e-06
UniRef100_Q5AED9 Branchpoint-bridging protein n=1 Tax=Candida al... 56 2e-06
UniRef100_B7PZF7 Putative uncharacterized protein (Fragment) n=1... 56 3e-06
UniRef100_B4M801 GJ16689 n=1 Tax=Drosophila virilis RepID=B4M801... 56 3e-06
UniRef100_B4GVT9 GL14605 n=1 Tax=Drosophila persimilis RepID=B4G... 56 3e-06
UniRef100_Q2HEQ9 Predicted protein n=1 Tax=Chaetomium globosum R... 56 3e-06
UniRef100_UPI0001926BC8 PREDICTED: similar to mini-collagen isof... 56 3e-06
UniRef100_UPI0001796E5C PREDICTED: similar to Formin-binding pro... 56 3e-06
UniRef100_UPI0000F2C630 PREDICTED: similar to zinc finger homeod... 56 3e-06
UniRef100_UPI0000F214B9 PREDICTED: similar to Cyclic nucleotide-... 56 3e-06
UniRef100_UPI000069F5FD Protein FAM54A (DUF729 domain-containing... 56 3e-06
UniRef100_UPI00004D5D58 Protein FAM54A (DUF729 domain-containing... 56 3e-06
UniRef100_UPI00016E7DED UPI00016E7DED related cluster n=1 Tax=Ta... 56 3e-06
UniRef100_UPI00016E72DF UPI00016E72DF related cluster n=1 Tax=Ta... 56 3e-06
UniRef100_UPI00005A1D18 PREDICTED: hypothetical protein XP_84743... 56 3e-06
UniRef100_Q6GQB0 MGC80202 protein n=1 Tax=Xenopus laevis RepID=Q... 56 3e-06
UniRef100_Q0P4U6 Putative uncharacterized protein MGC145636 n=1 ... 56 3e-06
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 56 3e-06
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 56 3e-06
UniRef100_A0ADW6 Putative secreted proline-rich protein n=1 Tax=... 56 3e-06
UniRef100_Q6EUQ1 Os02g0176100 protein n=1 Tax=Oryza sativa Japon... 56 3e-06
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 56 3e-06
UniRef100_Q00485 Mini-collagen (Fragment) n=1 Tax=Hydra sp. RepI... 56 3e-06
UniRef100_C0JIS9 Nematoblast-specific protein nb039a-sv9 (Fragme... 56 3e-06
UniRef100_Q2H4B7 Predicted protein n=1 Tax=Chaetomium globosum R... 56 3e-06
UniRef100_C5G899 Predicted protein n=1 Tax=Ajellomyces dermatiti... 56 3e-06
UniRef100_Q9AKJ0 Arp2/3 complex-activating protein rickA n=1 Tax... 56 3e-06
UniRef100_UPI0000D9E432 PREDICTED: similar to formin-like 1 isof... 55 3e-06
UniRef100_UPI0000D9E42C PREDICTED: similar to formin-like 1 isof... 55 3e-06
UniRef100_UPI0000D9D8AD PREDICTED: similar to formin binding pro... 55 3e-06
UniRef100_UPI0000D9D8AC PREDICTED: similar to formin binding pro... 55 3e-06
UniRef100_Q69ZN8 MKIAA1205 protein (Fragment) n=3 Tax=Mus muscul... 55 3e-06
UniRef100_UPI00016E10B6 UPI00016E10B6 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI00016E10A1 UPI00016E10A1 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 55 3e-06
UniRef100_C1DRF4 TolA protein n=1 Tax=Azotobacter vinelandii DJ ... 55 3e-06
UniRef100_B9JZN3 Putative uncharacterized protein n=1 Tax=Agroba... 55 3e-06
UniRef100_B0BYE9 Arp2/3 complex activation protein n=1 Tax=Ricke... 55 3e-06
UniRef100_C5ABQ4 Putative uncharacterized protein n=1 Tax=Burkho... 55 3e-06
UniRef100_A3WEW6 Peptidoglycan-associated protein n=1 Tax=Erythr... 55 3e-06
UniRef100_A3UDI3 OmpA family protein n=1 Tax=Oceanicaulis alexan... 55 3e-06
UniRef100_Q7XHB6 Transposon protein, putative, CACTA, En/Spm sub... 55 3e-06
UniRef100_C1MYV9 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 3e-06
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 55 3e-06
UniRef100_C0P373 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_B9RYC5 Serine-threonine protein kinase, plant-type, pu... 55 3e-06
UniRef100_B7FQ54 Predicted protein n=1 Tax=Phaeodactylum tricorn... 55 3e-06
UniRef100_B6U4Z9 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_B4F904 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_A4S9A6 Predicted protein n=1 Tax=Ostreococcus lucimari... 55 3e-06
UniRef100_Q9W363 CG12124 n=2 Tax=Drosophila melanogaster RepID=Q... 55 3e-06
UniRef100_Q5CS67 Signal peptide containing large protein with pr... 55 3e-06
UniRef100_Q581D1 Flagellum-adhesion glycoprotein, putative n=1 T... 55 3e-06
UniRef100_Q581C6 Flagellum-adhesion glycoprotein, putative n=1 T... 55 3e-06
UniRef100_Q4N8Y1 Putative uncharacterized protein n=1 Tax=Theile... 55 3e-06
UniRef100_Q4CRT2 Dispersed gene family protein 1 (DGF-1), putati... 55 3e-06
UniRef100_B4LCA6 GJ12883 n=1 Tax=Drosophila virilis RepID=B4LCA6... 55 3e-06
UniRef100_B2KTD4 Minicollagen 1 n=1 Tax=Clytia hemisphaerica Rep... 55 3e-06
UniRef100_C4JU13 Putative uncharacterized protein n=1 Tax=Uncino... 55 3e-06
UniRef100_C1G9Q7 Putative uncharacterized protein n=1 Tax=Paraco... 55 3e-06
UniRef100_C0S1D1 Putative uncharacterized protein n=1 Tax=Paraco... 55 3e-06
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 55 3e-06
UniRef100_UPI0001983B64 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0000DF063E Os02g0254800 n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_C7IY26 Os02g0254800 protein n=1 Tax=Oryza sativa Japon... 55 4e-06
UniRef100_B9F4U6 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_A4H5Q3 Cleavage and polyadenylation specificity factor... 55 4e-06
UniRef100_UPI000187E87F hypothetical protein MPER_09495 n=1 Tax=... 55 5e-06
UniRef100_UPI000180CA59 PREDICTED: similar to formin, inverted n... 55 5e-06
UniRef100_UPI000155D461 PREDICTED: similar to Mitogen-activated ... 55 5e-06
UniRef100_UPI0000E804FF PREDICTED: similar to Formin binding pro... 55 5e-06
UniRef100_UPI000059FC15 PREDICTED: similar to microtubule-associ... 55 5e-06
UniRef100_UPI0000D67D8C F-box protein 11 n=1 Tax=Mus musculus Re... 55 5e-06
UniRef100_UPI000013D567 Huntingtin (Huntington disease protein) ... 55 5e-06
UniRef100_UPI0000ECB7FD Formin-binding protein 4 (Formin-binding... 55 5e-06
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 55 5e-06
UniRef100_Q0ILB7 ORF1629 n=1 Tax=Leucania separata nuclear polyh... 55 5e-06
UniRef100_B8H5M9 Cell surface antigen Sca2 n=2 Tax=Caulobacter v... 55 5e-06
UniRef100_B3R5T8 Membrane spanning protein in TolA-TolQ-TolR com... 55 5e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 55 5e-06
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 55 5e-06
UniRef100_C5XWL6 Putative uncharacterized protein Sb04g037900 n=... 55 5e-06