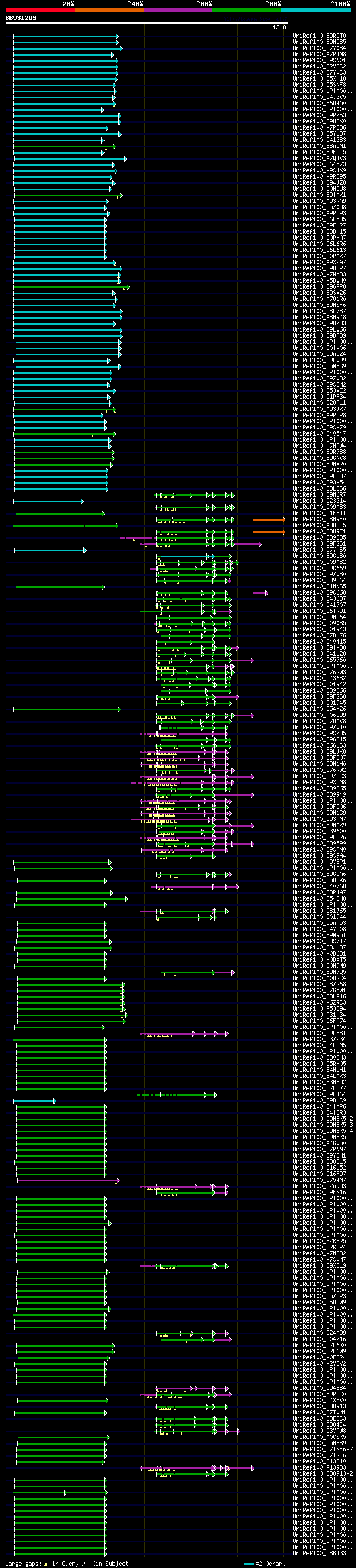
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB931203 RCC02975
(1218 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9RQT0 Serine/threonine-protein kinase, putative n=1 T... 266 3e-69
UniRef100_B9HDB5 Predicted protein n=1 Tax=Populus trichocarpa R... 262 6e-68
UniRef100_Q7Y0S4 Protein kinase n=1 Tax=Raphanus sativus RepID=Q... 261 1e-67
UniRef100_A7P4N8 Chromosome chr4 scaffold_6, whole genome shotgu... 258 7e-67
UniRef100_Q9SN01 Putative uncharacterized protein AT4g33080 n=1 ... 258 1e-66
UniRef100_Q2V3C2 Putative uncharacterized protein At4g33080.2 n=... 258 1e-66
UniRef100_Q7Y0S3 Protein kinase n=1 Tax=Raphanus sativus RepID=Q... 254 1e-65
UniRef100_C5XM10 Putative uncharacterized protein Sb03g003320 n=... 244 1e-62
UniRef100_Q5SNF8 Os01g0186700 protein n=1 Tax=Oryza sativa Japon... 243 3e-62
UniRef100_UPI0001984163 PREDICTED: hypothetical protein n=1 Tax=... 240 2e-61
UniRef100_C4J3V5 Putative uncharacterized protein n=1 Tax=Zea ma... 238 8e-61
UniRef100_B6U4A0 Serine/threonine-protein kinase 38 n=1 Tax=Zea ... 238 1e-60
UniRef100_UPI000198312D PREDICTED: similar to protein kinase n=1... 236 3e-60
UniRef100_B9RK53 Serine/threonine-protein kinase, putative n=1 T... 233 3e-59
UniRef100_B9HDX0 Predicted protein n=1 Tax=Populus trichocarpa R... 233 4e-59
UniRef100_A7PE36 Chromosome chr11 scaffold_13, whole genome shot... 229 4e-58
UniRef100_C5YU87 Putative uncharacterized protein Sb09g005720 n=... 226 4e-57
UniRef100_Q41383 Protein kinase n=1 Tax=Spinacia oleracea RepID=... 225 7e-57
UniRef100_B8ADN1 Putative uncharacterized protein n=1 Tax=Oryza ... 224 1e-56
UniRef100_B9ETJ5 Putative uncharacterized protein n=1 Tax=Oryza ... 221 2e-55
UniRef100_A7Q4V3 Chromosome chr10 scaffold_50, whole genome shot... 219 6e-55
UniRef100_O64573 Putative uncharacterized protein At2g19400 n=1 ... 218 8e-55
UniRef100_A9SJX9 Predicted protein n=1 Tax=Physcomitrella patens... 218 8e-55
UniRef100_A9RQ95 Predicted protein n=1 Tax=Physcomitrella patens... 217 2e-54
UniRef100_Q94JZ0 Putative uncharacterized protein At2g19400; F27... 217 2e-54
UniRef100_C0HGU8 Putative uncharacterized protein n=1 Tax=Zea ma... 216 3e-54
UniRef100_B9I0X1 Predicted protein n=1 Tax=Populus trichocarpa R... 215 9e-54
UniRef100_A9SKA9 Predicted protein n=1 Tax=Physcomitrella patens... 215 9e-54
UniRef100_C5Z0U8 Putative uncharacterized protein Sb09g025200 n=... 212 6e-53
UniRef100_A9RQ93 Predicted protein n=1 Tax=Physcomitrella patens... 212 6e-53
UniRef100_Q6L535 Os05g0511400 protein n=1 Tax=Oryza sativa Japon... 212 8e-53
UniRef100_B9FL27 Putative uncharacterized protein n=1 Tax=Oryza ... 212 8e-53
UniRef100_B8B015 Putative uncharacterized protein n=1 Tax=Oryza ... 212 8e-53
UniRef100_C0PHA7 Putative uncharacterized protein n=1 Tax=Zea ma... 211 1e-52
UniRef100_Q6L6R6 Protein kinase n=1 Tax=Triticum aestivum RepID=... 211 1e-52
UniRef100_Q6L613 WNdr1D-like protein kinase n=4 Tax=Triticeae Re... 211 1e-52
UniRef100_C0PAX7 Putative uncharacterized protein n=1 Tax=Zea ma... 211 1e-52
UniRef100_A9SKA7 Predicted protein n=1 Tax=Physcomitrella patens... 211 1e-52
UniRef100_B9H8P7 Predicted protein n=1 Tax=Populus trichocarpa R... 210 2e-52
UniRef100_A7NXD3 Chromosome chr5 scaffold_2, whole genome shotgu... 208 8e-52
UniRef100_A5BWH0 Putative uncharacterized protein n=1 Tax=Vitis ... 208 8e-52
UniRef100_B9GRP0 Predicted protein n=1 Tax=Populus trichocarpa R... 207 1e-51
UniRef100_B9SV26 Serine/threonine-protein kinase, putative n=1 T... 207 2e-51
UniRef100_A7Q1R0 Chromosome chr7 scaffold_44, whole genome shotg... 207 2e-51
UniRef100_B9HSF6 Predicted protein n=1 Tax=Populus trichocarpa R... 203 3e-50
UniRef100_Q8L7S7 AT4g14350/dl3215c n=1 Tax=Arabidopsis thaliana ... 202 5e-50
UniRef100_A8MR48 Uncharacterized protein At4g14350.3 n=1 Tax=Ara... 202 5e-50
UniRef100_B9HKH3 Predicted protein n=1 Tax=Populus trichocarpa R... 202 6e-50
UniRef100_Q9LW66 Protein kinase n=1 Tax=Arabidopsis thaliana Rep... 201 1e-49
UniRef100_B9DF89 AT3G23310 protein (Fragment) n=1 Tax=Arabidopsi... 201 1e-49
UniRef100_UPI0000DD99F6 Os10g0476100 n=1 Tax=Oryza sativa Japoni... 199 4e-49
UniRef100_Q0IX06 Os10g0476100 protein n=1 Tax=Oryza sativa Japon... 199 4e-49
UniRef100_Q9AUZ4 Putative uncharacterized protein n=3 Tax=Oryza ... 199 4e-49
UniRef100_Q9LW99 Protein kinase MK6 n=1 Tax=Mesembryanthemum cry... 199 5e-49
UniRef100_C5WYG9 Putative uncharacterized protein Sb01g019410 n=... 198 9e-49
UniRef100_UPI0000162C3F protein kinase, putative n=1 Tax=Arabido... 197 2e-48
UniRef100_Q9ZWB2 F21M11.15 protein n=1 Tax=Arabidopsis thaliana ... 197 2e-48
UniRef100_Q9SIM2 Putative uncharacterized protein At2g20470 n=1 ... 197 3e-48
UniRef100_Q53VE2 Ser/Thr protein kinase n=1 Tax=Lotus japonicus ... 197 3e-48
UniRef100_Q1PF34 Protein kinase n=1 Tax=Arabidopsis thaliana Rep... 197 3e-48
UniRef100_Q2QTL1 Putative uncharacterized protein n=2 Tax=Oryza ... 196 4e-48
UniRef100_A9SJX7 Predicted protein n=1 Tax=Physcomitrella patens... 195 7e-48
UniRef100_A9RIR8 Predicted protein n=1 Tax=Physcomitrella patens... 194 2e-47
UniRef100_UPI0000162F2A protein kinase, putative n=1 Tax=Arabido... 191 2e-46
UniRef100_Q9SA79 T5I8.9 protein n=1 Tax=Arabidopsis thaliana Rep... 191 2e-46
UniRef100_Q40547 Protein kinase n=1 Tax=Nicotiana tabacum RepID=... 187 3e-45
UniRef100_UPI000198546F PREDICTED: hypothetical protein n=1 Tax=... 183 4e-44
UniRef100_A7NTW4 Chromosome chr18 scaffold_1, whole genome shotg... 183 4e-44
UniRef100_B9R7B8 Serine/threonine-protein kinase, putative n=1 T... 179 7e-43
UniRef100_B9GNV8 Predicted protein n=1 Tax=Populus trichocarpa R... 179 7e-43
UniRef100_B9MVR0 Predicted protein n=1 Tax=Populus trichocarpa R... 176 4e-42
UniRef100_UPI00005DC0F2 protein kinase, putative n=1 Tax=Arabido... 169 6e-40
UniRef100_Q9FIB7 Protein kinase n=1 Tax=Arabidopsis thaliana Rep... 169 6e-40
UniRef100_Q93V54 Ndr kinase n=1 Tax=Arabidopsis thaliana RepID=Q... 169 6e-40
UniRef100_Q8LDG6 Protein kinase n=1 Tax=Arabidopsis thaliana Rep... 169 6e-40
UniRef100_Q9M6R7 Extensin n=1 Tax=Pisum sativum RepID=Q9M6R7_PEA 157 2e-36
UniRef100_O23314 Protein kinase n=1 Tax=Arabidopsis thaliana Rep... 155 1e-35
UniRef100_Q09083 Hydroxyproline-rich glycoprotein n=1 Tax=Phaseo... 149 6e-34
UniRef100_C1EHI1 Predicted protein n=1 Tax=Micromonas sp. RCC299... 146 5e-33
UniRef100_Q8H9E0 Hydroxyproline-rich glycoprotein (Fragment) n=1... 145 9e-33
UniRef100_A8HQF5 Serine/threonine protein kinase 19 n=1 Tax=Chla... 143 3e-32
UniRef100_Q8H9E1 Extensin (Fragment) n=1 Tax=Solanum tuberosum R... 142 1e-31
UniRef100_Q39835 Extensin n=1 Tax=Glycine max RepID=Q39835_SOYBN 142 1e-31
UniRef100_Q9FSG1 Putative extensin n=1 Tax=Nicotiana sylvestris ... 140 4e-31
UniRef100_Q7Y0S5 Protein kinase n=1 Tax=Raphanus sativus RepID=Q... 139 6e-31
UniRef100_B9GU80 Predicted protein (Fragment) n=1 Tax=Populus tr... 138 1e-30
UniRef100_Q09082 Extensin (Class I) n=1 Tax=Solanum lycopersicum... 137 2e-30
UniRef100_Q9C669 Putative uncharacterized protein F28B23.9 n=1 T... 137 3e-30
UniRef100_Q9ZW80 Putative extensin n=1 Tax=Arabidopsis thaliana ... 136 4e-30
UniRef100_Q39864 Hydroxyproline-rich glycoprotein (Fragment) n=1... 136 5e-30
UniRef100_C1MNG5 Predicted protein n=1 Tax=Micromonas pusilla CC... 135 7e-30
UniRef100_Q9C668 Putative uncharacterized protein F28B23.10 n=1 ... 134 2e-29
UniRef100_Q43687 Extensin-like protein (Fragment) n=1 Tax=Vigna ... 134 2e-29
UniRef100_Q41707 Extensin class 1 protein n=1 Tax=Vigna unguicul... 134 2e-29
UniRef100_C6TK91 Putative uncharacterized protein n=1 Tax=Glycin... 134 2e-29
UniRef100_Q9M564 CRANTZ hydroxyproline-rich glycoprotein n=1 Tax... 134 3e-29
UniRef100_Q09085 Hydroxyproline-rich glycoprotein (HRGP) (Fragme... 134 3e-29
UniRef100_Q01943 Extensin (Class I) (Fragment) n=1 Tax=Solanum l... 133 3e-29
UniRef100_Q7DLZ6 Extensin-like protein (Fragment) n=1 Tax=Vigna ... 132 8e-29
UniRef100_Q40415 Extensin (Fragment) n=1 Tax=Nicotiana sylvestri... 132 8e-29
UniRef100_B9IAD8 Predicted protein n=1 Tax=Populus trichocarpa R... 131 1e-28
UniRef100_Q41120 Hydroxyproline-rich glycoprotein (HRGP) (Fragme... 131 2e-28
UniRef100_O65760 Extensin (Fragment) n=1 Tax=Cicer arietinum Rep... 130 2e-28
UniRef100_UPI00019859A1 PREDICTED: hypothetical protein n=1 Tax=... 130 4e-28
UniRef100_Q76KW3 Hydroxyproline-rich glycoprotein-2 (Fragment) n... 130 4e-28
UniRef100_Q43682 Extensin-like protein (Fragment) n=1 Tax=Vigna ... 130 4e-28
UniRef100_Q01942 Extensin (Class I) n=1 Tax=Solanum lycopersicum... 130 4e-28
UniRef100_Q39866 Hydroxyproline-rich glycoprotein (Fragment) n=1... 129 7e-28
UniRef100_Q9FSG0 Extensin n=1 Tax=Nicotiana sylvestris RepID=Q9F... 129 9e-28
UniRef100_Q01945 Extensin (Class I) (Fragment) n=1 Tax=Solanum l... 129 9e-28
UniRef100_Q54Y26 Probable serine/threonine-protein kinase ndrA n... 129 9e-28
UniRef100_P06599 Extensin n=1 Tax=Daucus carota RepID=EXTN_DAUCA 129 9e-28
UniRef100_Q7DMV8 Hydroxyproline-rich glycoprotein (HRGP) (Fragme... 128 1e-27
UniRef100_Q9ZWT0 Extensin n=1 Tax=Adiantum capillus-veneris RepI... 127 2e-27
UniRef100_Q9SK35 Putative uncharacterized protein At2g24980 n=1 ... 127 3e-27
UniRef100_B9GF15 Predicted protein n=1 Tax=Populus trichocarpa R... 127 3e-27
UniRef100_Q6GUG3 Serine/proline-rich repeat protein n=2 Tax=Lupi... 126 4e-27
UniRef100_Q9LJK0 Genomic DNA, chromosome 3, P1 clone: MZN14 n=1 ... 126 6e-27
UniRef100_Q9FG07 Gb|AAD23015.1 n=2 Tax=Arabidopsis thaliana RepI... 125 7e-27
UniRef100_Q9M1H0 Extensin-like protein n=1 Tax=Arabidopsis thali... 125 7e-27
UniRef100_Q76KW2 Hydroxyproline-rich glycoprotein-3 (Fragment) n... 125 9e-27
UniRef100_Q9ZUC3 F5O8.27 protein n=1 Tax=Arabidopsis thaliana Re... 125 1e-26
UniRef100_Q9STM8 Extensin-like protein n=1 Tax=Arabidopsis thali... 125 1e-26
UniRef100_Q39865 Hydroxyproline-rich glycoprotein (Fragment) n=1... 124 2e-26
UniRef100_Q39949 Hydroxyproline-rich protein n=1 Tax=Helianthus ... 124 2e-26
UniRef100_UPI0001739340 ATHRGP1 (HYDROXYPROLINE-RICH GLYCOPROTEI... 124 3e-26
UniRef100_Q9FG06 Genomic DNA, chromosome 5, BAC clone:F15M7 n=1 ... 124 3e-26
UniRef100_Q9M1G9 Extensin-2 n=1 Tax=Arabidopsis thaliana RepID=E... 124 3e-26
UniRef100_Q9STM7 Extensin-like protein n=1 Tax=Arabidopsis thali... 123 4e-26
UniRef100_B9NAX9 Predicted protein n=1 Tax=Populus trichocarpa R... 123 4e-26
UniRef100_Q39600 Extensin n=1 Tax=Catharanthus roseus RepID=Q396... 123 5e-26
UniRef100_Q9FH26 Genomic DNA, chromosome 5, TAC clone: K20J1 n=1... 122 6e-26
UniRef100_Q39599 Extensin n=1 Tax=Catharanthus roseus RepID=Q395... 122 8e-26
UniRef100_Q9STN0 Extensin-like protein n=1 Tax=Arabidopsis thali... 122 1e-25
UniRef100_Q9S9A4 Extensin (Fragment) n=1 Tax=Vicia faba RepID=Q9... 122 1e-25
UniRef100_A9V8P1 Predicted protein n=1 Tax=Monosiga brevicollis ... 121 2e-25
UniRef100_UPI00016E01EF UPI00016E01EF related cluster n=2 Tax=Ta... 120 3e-25
UniRef100_B9GWA6 Predicted protein (Fragment) n=1 Tax=Populus tr... 120 4e-25
UniRef100_C5DZK6 ZYRO0G05214p n=1 Tax=Zygosaccharomyces rouxii C... 120 4e-25
UniRef100_Q40768 Extensin n=1 Tax=Prunus dulcis RepID=Q40768_PRUDU 119 5e-25
UniRef100_B3RJA7 Putative uncharacterized protein n=1 Tax=Tricho... 119 5e-25
UniRef100_Q54IH8 Probable serine/threonine-protein kinase ndrB n... 119 5e-25
UniRef100_UPI00017B0E12 UPI00017B0E12 related cluster n=1 Tax=Te... 119 7e-25
UniRef100_O81765 Extensin-like protein n=1 Tax=Arabidopsis thali... 119 7e-25
UniRef100_Q01944 Extensin (Class I) (Fragment) n=1 Tax=Solanum l... 119 9e-25
UniRef100_Q5AP53 Likely protein kinase n=1 Tax=Candida albicans ... 119 9e-25
UniRef100_C4YD08 Serine/threonine-protein kinase CBK1 n=1 Tax=Ca... 119 9e-25
UniRef100_B9W951 Serine/threonine protein kinase, putative (Cell... 119 9e-25
UniRef100_C3S7I7 Serine/threonine protein kinase 38 n=1 Tax=Cras... 118 1e-24
UniRef100_B8JM87 Novel protein similar to H.sapien STK38, serine... 117 2e-24
UniRef100_A0D631 Chromosome undetermined scaffold_39, whole geno... 117 3e-24
UniRef100_A0BXT5 Chromosome undetermined scaffold_135, whole gen... 117 3e-24
UniRef100_C0H9M9 Serine/threonine-protein kinase 38 n=1 Tax=Salm... 117 3e-24
UniRef100_B9H7Q5 Predicted protein n=1 Tax=Populus trichocarpa R... 117 3e-24
UniRef100_A0DKC4 Chromosome undetermined scaffold_54, whole geno... 117 3e-24
UniRef100_C8ZG68 Cbk1p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 117 3e-24
UniRef100_C7GXW1 Cbk1p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 117 3e-24
UniRef100_B3LP16 Serine/threonine protein kinase n=1 Tax=Sacchar... 117 3e-24
UniRef100_A6ZRS3 Serine/threonine protein kinase n=1 Tax=Sacchar... 117 3e-24
UniRef100_P53894 Serine/threonine-protein kinase CBK1 n=1 Tax=Sa... 117 3e-24
UniRef100_P31034 Serine/threonine-protein kinase CBK1 n=1 Tax=Kl... 116 4e-24
UniRef100_Q6FP74 Serine/threonine-protein kinase CBK1 n=1 Tax=Ca... 116 4e-24
UniRef100_UPI00016E01EE UPI00016E01EE related cluster n=1 Tax=Ta... 116 6e-24
UniRef100_Q9LHS1 Genomic DNA, chromosome 5, TAC clone:K3D20 n=2 ... 116 6e-24
UniRef100_C3ZK34 Putative uncharacterized protein n=1 Tax=Branch... 116 6e-24
UniRef100_B4LBM5 GJ14000 n=1 Tax=Drosophila virilis RepID=B4LBM5... 115 7e-24
UniRef100_UPI0001B7B937 serine/threonine kinase 38 like n=1 Tax=... 115 1e-23
UniRef100_Q803H3 Serine/threonine kinase 38 like n=1 Tax=Danio r... 115 1e-23
UniRef100_Q5RH05 Serine/threonine kinase 38 like n=1 Tax=Danio r... 115 1e-23
UniRef100_B4MLH1 GK17215 n=1 Tax=Drosophila willistoni RepID=B4M... 115 1e-23
UniRef100_B4L0X3 GI13665 n=1 Tax=Drosophila mojavensis RepID=B4L... 115 1e-23
UniRef100_B3M8U2 GF10844 n=1 Tax=Drosophila ananassae RepID=B3M8... 115 1e-23
UniRef100_Q2LZZ7 Serine/threonine-protein kinase 38-like n=2 Tax... 115 1e-23
UniRef100_Q9LJ64 Extensin protein-like n=1 Tax=Arabidopsis thali... 114 2e-23
UniRef100_B9DHS9 AT4G33080 protein (Fragment) n=2 Tax=Arabidopsi... 114 2e-23
UniRef100_B4IXP6 GH16909 n=1 Tax=Drosophila grimshawi RepID=B4IX... 114 2e-23
UniRef100_B4IIR3 GM16004 n=1 Tax=Drosophila sechellia RepID=B4II... 114 2e-23
UniRef100_Q9NBK5-2 Isoform A of Serine/threonine-protein kinase ... 114 2e-23
UniRef100_Q9NBK5-3 Isoform B of Serine/threonine-protein kinase ... 114 2e-23
UniRef100_Q9NBK5-4 Isoform C of Serine/threonine-protein kinase ... 114 2e-23
UniRef100_Q9NBK5 Serine/threonine-protein kinase 38-like n=1 Tax... 114 2e-23
UniRef100_A4GW50 Serine/threonine kinase 38 like n=1 Tax=Rattus ... 114 2e-23
UniRef100_Q7PNN7 AGAP005521-PA n=1 Tax=Anopheles gambiae RepID=Q... 114 2e-23
UniRef100_Q9Y2H1 Serine/threonine-protein kinase 38-like n=2 Tax... 114 2e-23
UniRef100_Q803L5 Novel protein similar to H.sapiens STK38, serin... 114 3e-23
UniRef100_Q16U52 Serine/threonine-protein kinase 38 (Ndr2 protei... 114 3e-23
UniRef100_Q16F97 Serine/threonine-protein kinase 38 (Ndr2 protei... 114 3e-23
UniRef100_Q754N7 Serine/threonine-protein kinase CBK1 n=1 Tax=Er... 114 3e-23
UniRef100_Q2A9D3 Extensin, putative n=1 Tax=Brassica oleracea Re... 113 4e-23
UniRef100_Q9FS16 Extensin-3 n=1 Tax=Arabidopsis thaliana RepID=E... 113 4e-23
UniRef100_UPI00017962AF PREDICTED: similar to serine/threonine k... 113 5e-23
UniRef100_UPI000155C0AC PREDICTED: similar to Serine/threonine k... 113 5e-23
UniRef100_UPI0000F2E48C PREDICTED: similar to Serine/threonine k... 113 5e-23
UniRef100_UPI0000D9CBB1 PREDICTED: similar to serine/threonine k... 113 5e-23
UniRef100_UPI0000D55BCF PREDICTED: similar to serine/threonine-p... 113 5e-23
UniRef100_UPI00005A4D0D PREDICTED: similar to serine/threonine k... 113 5e-23
UniRef100_UPI00016E6BD2 UPI00016E6BD2 related cluster n=1 Tax=Ta... 113 5e-23
UniRef100_B2KFR5 Serine/threonine kinase 38 like n=1 Tax=Mus mus... 113 5e-23
UniRef100_B2KFR4 Serine/threonine kinase 38 like n=1 Tax=Mus mus... 113 5e-23
UniRef100_A7MB32 STK38L protein n=1 Tax=Bos taurus RepID=A7MB32_... 113 5e-23
UniRef100_A7S0M7 Predicted protein n=1 Tax=Nematostella vectensi... 113 5e-23
UniRef100_Q9XIL9 Putative uncharacterized protein At2g15880 n=1 ... 112 6e-23
UniRef100_UPI00006CCA54 Protein kinase domain containing protein... 112 8e-23
UniRef100_UPI0000ECD256 serine/threonine kinase 38 like n=1 Tax=... 112 8e-23
UniRef100_UPI0000ECD255 serine/threonine kinase 38 like n=1 Tax=... 112 8e-23
UniRef100_UPI000060E2D2 serine/threonine kinase 38 like n=1 Tax=... 112 8e-23
UniRef100_Q5ZLR3 Putative uncharacterized protein n=1 Tax=Gallus... 112 8e-23
UniRef100_C5DCW9 KLTH0B06424p n=1 Tax=Lachancea thermotolerans C... 112 8e-23
UniRef100_UPI0000DB717F PREDICTED: similar to tricornered CG8637... 112 1e-22
UniRef100_UPI00006A5432 PREDICTED: similar to serine/threonine k... 111 1e-22
UniRef100_UPI000179D0BF UPI000179D0BF related cluster n=1 Tax=Bo... 111 1e-22
UniRef100_UPI0000F309D3 UPI0000F309D3 related cluster n=1 Tax=Bo... 111 1e-22
UniRef100_O24099 MtN12 protein (Fragment) n=1 Tax=Medicago trunc... 111 1e-22
UniRef100_O04216 Extensin (Fragment) n=1 Tax=Bromheadia finlayso... 111 1e-22
UniRef100_Q2L6X0 Sensory axon guidance protein 1, isoform b n=1 ... 111 1e-22
UniRef100_Q2L6W9 Sensory axon guidance protein 1, isoform a n=2 ... 111 1e-22
UniRef100_A0ED24 Chromosome undetermined scaffold_9, whole genom... 111 1e-22
UniRef100_A2VDV2 Serine/threonine-protein kinase 38 n=1 Tax=Bos ... 111 1e-22
UniRef100_UPI0001791BED PREDICTED: similar to serine/threonine-p... 111 2e-22
UniRef100_UPI0001791BEC PREDICTED: similar to serine/threonine-p... 111 2e-22
UniRef100_UPI0001791BEB PREDICTED: similar to serine/threonine-p... 111 2e-22
UniRef100_Q94ES4 Root nodule extensin (Fragment) n=1 Tax=Pisum s... 111 2e-22
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 111 2e-22
UniRef100_C4XYV0 Putative uncharacterized protein n=1 Tax=Clavis... 111 2e-22
UniRef100_Q38913 Extensin-1 n=1 Tax=Arabidopsis thaliana RepID=E... 111 2e-22
UniRef100_Q7T0M1 Trc-prov protein n=1 Tax=Xenopus laevis RepID=Q... 110 2e-22
UniRef100_Q3ECC3 Putative uncharacterized protein At1g76930.1 n=... 110 2e-22
UniRef100_Q304C4 Putative uncharacterized protein At1g76930.2 n=... 110 2e-22
UniRef100_C3VPW8 Extensin n=1 Tax=Lithospermum erythrorhizon Rep... 110 2e-22
UniRef100_A0CSK5 Chromosome undetermined scaffold_26, whole geno... 110 2e-22
UniRef100_C5MB89 Serine/threonine-protein kinase CBK1 n=1 Tax=Ca... 110 2e-22
UniRef100_Q7TSE6-2 Isoform 2 of Serine/threonine-protein kinase ... 110 2e-22
UniRef100_Q7TSE6 Serine/threonine-protein kinase 38-like n=1 Tax... 110 2e-22
UniRef100_O13310 Serine/threonine-protein kinase orb6 n=1 Tax=Sc... 110 2e-22
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 110 2e-22
UniRef100_Q38913-2 Isoform 2 of Extensin-1 n=1 Tax=Arabidopsis t... 110 2e-22
UniRef100_UPI00017EF8DA PREDICTED: similar to Serine/threonine-p... 110 3e-22
UniRef100_UPI00017975D6 PREDICTED: similar to Serine/threonine-p... 110 3e-22
UniRef100_UPI0000E4957D PREDICTED: similar to serine/threonine-p... 110 3e-22
UniRef100_UPI0000E20F31 PREDICTED: serine/threonine kinase 38 n=... 110 3e-22
UniRef100_UPI0000E20F30 PREDICTED: serine/threonine kinase 38 n=... 110 3e-22
UniRef100_UPI0000D9AC96 PREDICTED: serine/threonine kinase 38 is... 110 3e-22
UniRef100_UPI00005EB1AE PREDICTED: similar to GDP dissociation i... 110 3e-22
UniRef100_UPI00005A26A3 PREDICTED: similar to Serine/threonine-p... 110 3e-22
UniRef100_UPI000036D718 PREDICTED: serine/threonine kinase 38 n=... 110 3e-22
UniRef100_UPI00006A0732 Serine/threonine-protein kinase 38 (EC 2... 110 3e-22
UniRef100_UPI000017E1ED serine/threonine kinase 38 n=1 Tax=Rattu... 110 3e-22
UniRef100_UPI00004BBBAD Serine/threonine-protein kinase 38 (EC 2... 110 3e-22
UniRef100_Q8BJ33 Putative uncharacterized protein n=1 Tax=Mus mu... 110 3e-22
UniRef100_Q40793 Tyrosine-rich hydroxyproline-rich glycoprotein ... 110 3e-22
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 110 3e-22
UniRef100_Q5R6L0 Putative uncharacterized protein DKFZp469F104 n... 110 3e-22
UniRef100_B7PIM9 Serine/threonine protein kinase, putative n=1 T... 110 3e-22
UniRef100_A0DL93 Chromosome undetermined scaffold_55, whole geno... 110 3e-22
UniRef100_B2R6F9 cDNA, FLJ92932, highly similar to Homo sapiens ... 110 3e-22
UniRef100_C4R3T8 Serine/threonine protein kinase n=1 Tax=Pichia ... 110 3e-22
UniRef100_A3LQA3 Serine/threonine-protein kinase CBK1 (Fragment)... 110 3e-22
UniRef100_Q91VJ4 Serine/threonine-protein kinase 38 n=2 Tax=Mus ... 110 3e-22
UniRef100_Q15208 Serine/threonine-protein kinase 38 n=2 Tax=Homi... 110 3e-22
UniRef100_UPI000179F182 UPI000179F182 related cluster n=1 Tax=Bo... 110 4e-22
UniRef100_Q2A9D1 Extensin-like region containing protein n=1 Tax... 110 4e-22
UniRef100_Q5KEJ1 Serine/threonine-protein kinase orb6, putative ... 110 4e-22
UniRef100_Q55NX0 Putative uncharacterized protein n=1 Tax=Filoba... 110 4e-22
UniRef100_Q9STN1 Extensin-like protein n=1 Tax=Arabidopsis thali... 109 5e-22
UniRef100_A0BHF2 Chromosome undetermined scaffold_108, whole gen... 109 5e-22
UniRef100_Q9T0L0 Extensin-like protein n=1 Tax=Arabidopsis thali... 109 7e-22
UniRef100_UPI0000EB0B87 Serine/threonine-protein kinase 38-like ... 109 7e-22
UniRef100_Q6DIL2 Serine/threonine kinase 38 like n=1 Tax=Xenopus... 109 7e-22
UniRef100_A8XJL7 C. briggsae CBR-SAX-1 protein n=2 Tax=Caenorhab... 109 7e-22
UniRef100_Q9UPD3 Ndr Ser/Thr kinase-like protein (Fragment) n=1 ... 109 7e-22
UniRef100_B0CYD4 Predicted protein (Fragment) n=1 Tax=Laccaria b... 109 7e-22
UniRef100_UPI000194DDA4 PREDICTED: serine/threonine kinase 38 n=... 108 9e-22
UniRef100_UPI0000ECA0EA Serine/threonine-protein kinase 38 (EC 2... 108 9e-22
UniRef100_UPI00017B5049 UPI00017B5049 related cluster n=1 Tax=Te... 108 9e-22
UniRef100_Q94ES9 Root nodule extensin n=1 Tax=Pisum sativum RepI... 108 9e-22
UniRef100_Q94ES6 Root nodule extensin n=1 Tax=Pisum sativum RepI... 108 9e-22
UniRef100_Q94ES5 Root nodule extensin n=1 Tax=Pisum sativum RepI... 108 9e-22
UniRef100_B1N2K6 Protein kinase, putative n=1 Tax=Entamoeba hist... 108 9e-22
UniRef100_B6JW89 Serine/threonine-protein kinase orb6 n=1 Tax=Sc... 108 9e-22
UniRef100_O65375 F12F1.9 protein n=1 Tax=Arabidopsis thaliana Re... 108 1e-21
UniRef100_A8N678 Putative uncharacterized protein n=1 Tax=Coprin... 108 1e-21
UniRef100_A5DCA8 Putative uncharacterized protein n=1 Tax=Pichia... 108 1e-21
UniRef100_UPI00003BE20C hypothetical protein DEHA0F13959g n=1 Ta... 108 2e-21
UniRef100_Q6BLJ9 Serine/threonine-protein kinase CBK1 n=1 Tax=De... 108 2e-21
UniRef100_UPI000151A9E0 hypothetical protein PGUG_00913 n=1 Tax=... 107 2e-21
UniRef100_Q8LK15 Extensin n=1 Tax=Brassica napus RepID=Q8LK15_BRANA 107 2e-21
UniRef100_Q4PAX2 Putative uncharacterized protein n=1 Tax=Ustila... 107 2e-21
UniRef100_Q6NTL3 MGC83214 protein n=1 Tax=Xenopus laevis RepID=Q... 107 3e-21
UniRef100_B9IKD0 Predicted protein n=1 Tax=Populus trichocarpa R... 107 3e-21
UniRef100_B0EAW9 Serine/threonine protein kinase, putative n=1 T... 107 3e-21
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 107 3e-21
UniRef100_Q94ES3 Root nodule extensin (Fragment) n=1 Tax=Pisum s... 107 3e-21
UniRef100_C4QPL6 Serine/threonine kinase n=1 Tax=Schistosoma man... 107 3e-21
UniRef100_A0BCQ1 Chromosome undetermined scaffold_10, whole geno... 107 3e-21
UniRef100_Q6TGC6 Serine/threonine-protein kinase CBK1 n=1 Tax=Pn... 107 3e-21
UniRef100_B9RIB5 Extensin, proline-rich protein, putative n=1 Ta... 106 5e-21
UniRef100_A0EHT7 Chromosome undetermined scaffold_98, whole geno... 106 5e-21
UniRef100_A0DPF6 Chromosome undetermined scaffold_59, whole geno... 106 5e-21
UniRef100_A5DTJ0 Serine/threonine-protein kinase CBK1 n=1 Tax=Lo... 106 5e-21
UniRef100_Q9SPM1 Extensin-like protein n=1 Tax=Solanum lycopersi... 106 6e-21
UniRef100_Q2A9D2 Extensin-like region containing protein n=1 Tax... 106 6e-21
UniRef100_B9N4U0 Predicted protein (Fragment) n=1 Tax=Populus tr... 106 6e-21
UniRef100_A0DVE0 Chromosome undetermined scaffold_65, whole geno... 106 6e-21
UniRef100_Q01946 Extensin (Class I) n=1 Tax=Solanum lycopersicum... 105 8e-21
UniRef100_Q27145 Protein kinase (Fragment) n=1 Tax=Moneuplotes c... 105 8e-21
UniRef100_Q27141 Protein kinase n=1 Tax=Moneuplotes crassus RepI... 105 8e-21
UniRef100_A0EDW8 Chromosome undetermined scaffold_90, whole geno... 105 8e-21
UniRef100_A0E3S6 Chromosome undetermined scaffold_77, whole geno... 105 8e-21
UniRef100_A0C9E5 Chromosome undetermined scaffold_16, whole geno... 105 8e-21
UniRef100_B0D0X1 Predicted protein (Fragment) n=1 Tax=Laccaria b... 105 8e-21
UniRef100_Q5DC06 SJCHGC04903 protein n=1 Tax=Schistosoma japonic... 105 1e-20
UniRef100_B9SSV0 Cleavage and polyadenylation specificity factor... 105 1e-20
UniRef100_B9HRV2 Predicted protein n=1 Tax=Populus trichocarpa R... 105 1e-20
UniRef100_A8PX15 Putative uncharacterized protein n=1 Tax=Malass... 104 2e-20
UniRef100_UPI000186EDF6 serine/threonine-protein kinase, putativ... 104 2e-20
UniRef100_Q9XIB6 F13F21.7 protein n=1 Tax=Arabidopsis thaliana R... 103 3e-20
UniRef100_A0DGH3 Chromosome undetermined scaffold_5, whole genom... 103 3e-20
UniRef100_Q94ES8 Root nodule extensin (Fragment) n=1 Tax=Pisum s... 103 4e-20
UniRef100_Q06446 Extensin n=1 Tax=Solanum tuberosum RepID=Q06446... 101 6e-20
UniRef100_Q8RWX5 Putative uncharacterized protein At3g19020 (Fra... 102 7e-20
UniRef100_Q40402 Extensin (ext) n=1 Tax=Nicotiana plumbaginifoli... 102 7e-20
UniRef100_A8PLS0 Protein kinase domain containing protein n=1 Ta... 102 7e-20
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 102 9e-20
UniRef100_Q40503 Extensin n=1 Tax=Nicotiana tabacum RepID=Q40503... 102 9e-20
UniRef100_B9FLI1 Putative uncharacterized protein n=1 Tax=Oryza ... 102 9e-20
UniRef100_A2Y786 Putative uncharacterized protein n=1 Tax=Oryza ... 102 9e-20
UniRef100_Q4P4K7 Putative uncharacterized protein n=1 Tax=Ustila... 102 9e-20
UniRef100_O59918 Protein kinase Ukc1p (Fragment) n=1 Tax=Ustilag... 102 9e-20
UniRef100_Q40502 Extensin n=1 Tax=Nicotiana tabacum RepID=Q40502... 102 1e-19
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 101 2e-19
UniRef100_Q06802 Extensin n=1 Tax=Nicotiana tabacum RepID=Q06802... 101 2e-19
UniRef100_A3KD20 Leucine-rich repeat/extensin n=1 Tax=Nicotiana ... 101 2e-19
UniRef100_B0EAH2 Serine/threonine protein kinase, putative n=1 T... 101 2e-19
UniRef100_UPI0001A7B0C1 protein binding / structural constituent... 100 3e-19
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 100 3e-19
UniRef100_UPI000180B8AB PREDICTED: similar to GK13110 n=1 Tax=Ci... 100 3e-19
UniRef100_Q9SN46 Extensin-like protein n=1 Tax=Arabidopsis thali... 100 3e-19
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 100 3e-19
UniRef100_UPI000192550C PREDICTED: similar to mCG22338 n=1 Tax=H... 100 4e-19
UniRef100_Q94ES2 Root nodule extensin (Fragment) n=1 Tax=Pisum s... 100 4e-19
UniRef100_C6T6W7 Putative uncharacterized protein (Fragment) n=1... 100 6e-19
UniRef100_A9SB04 Predicted protein n=1 Tax=Physcomitrella patens... 100 6e-19
UniRef100_A4S2A2 Predicted protein n=1 Tax=Ostreococcus lucimari... 100 6e-19
UniRef100_C4MBB2 Protein kinase, putative n=1 Tax=Entamoeba hist... 99 7e-19
UniRef100_C4JS82 Serine/threonine-protein kinase cot-1 n=1 Tax=U... 99 7e-19
UniRef100_UPI0000E125D3 Os05g0552600 n=1 Tax=Oryza sativa Japoni... 99 9e-19
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 99 9e-19
UniRef100_A3KD22 Leucine-rich repeat/extensin 1 n=1 Tax=Nicotian... 99 9e-19
UniRef100_Q41983 Extensin precusor (Fragment) n=1 Tax=Arabidopsi... 99 1e-18
UniRef100_Q41400 Root nodule extensin (Fragment) n=1 Tax=Sesbani... 98 2e-18
UniRef100_B0CTV3 Predicted protein (Fragment) n=1 Tax=Laccaria b... 98 2e-18
UniRef100_Q09084 Extensin (Class II) n=1 Tax=Solanum lycopersicu... 98 2e-18
UniRef100_Q4PSF3 Proline-rich extensin-like family protein n=1 T... 97 3e-18
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 97 4e-18
UniRef100_Q94ES7 Root nodule extensin (Fragment) n=1 Tax=Pisum s... 97 5e-18
UniRef100_UPI00001631D5 LRX2 (LEUCINE-RICH REPEAT/EXTENSIN 2); p... 96 6e-18
UniRef100_Q43504 Extensin-like protein Dif10 (Fragment) n=1 Tax=... 96 6e-18
UniRef100_O48809 F24O1.18 n=1 Tax=Arabidopsis thaliana RepID=O48... 96 6e-18
UniRef100_B9MVI9 Predicted protein (Fragment) n=1 Tax=Populus tr... 96 6e-18
UniRef100_A2Q9A3 Similarity to serine/threonine kinase cot-1 - N... 96 6e-18
UniRef100_Q40150 Tomato cell wall HRGP (hydroxproline-rich glyco... 96 8e-18
UniRef100_C1GW62 Serine/threonine-protein kinase cot-1 n=1 Tax=P... 96 8e-18
UniRef100_UPI00017B1463 UPI00017B1463 related cluster n=1 Tax=Te... 96 1e-17
UniRef100_Q4RF98 Chromosome 14 SCAF15120, whole genome shotgun s... 96 1e-17
UniRef100_Q29CB7 GA11375 n=1 Tax=Drosophila pseudoobscura pseudo... 96 1e-17
UniRef100_B4GNB6 GL13988 n=1 Tax=Drosophila persimilis RepID=B4G... 96 1e-17
UniRef100_Q1E759 Putative uncharacterized protein n=1 Tax=Coccid... 96 1e-17
UniRef100_C5PG47 Serine/threonine-protein kinase, putative n=1 T... 96 1e-17
UniRef100_A8NXJ9 Putative uncharacterized protein n=1 Tax=Coprin... 96 1e-17
UniRef100_UPI0000E46629 PREDICTED: hypothetical protein, partial... 95 1e-17
UniRef100_B8LWD4 Serine/threonine protein kinase, putative n=1 T... 95 1e-17
UniRef100_UPI0001A2D0DA UPI0001A2D0DA related cluster n=1 Tax=Da... 95 2e-17
UniRef100_UPI0001A2D0D9 UPI0001A2D0D9 related cluster n=1 Tax=Da... 95 2e-17
UniRef100_B2BJC1 Lats2-1 n=1 Tax=Danio rerio RepID=B2BJC1_DANRE 95 2e-17
UniRef100_B2BJC0 Lats2-2 n=1 Tax=Danio rerio RepID=B2BJC0_DANRE 95 2e-17
UniRef100_B0S8I4 Novel protein similar to vertebrate LATS, large... 95 2e-17
UniRef100_Q76KW4 Hydroxyproline-rich glycoprotein-1 (Fragment) n... 95 2e-17
UniRef100_B9ILJ5 Predicted protein n=1 Tax=Populus trichocarpa R... 95 2e-17
UniRef100_B4PML7 GE23359 n=1 Tax=Drosophila yakuba RepID=B4PML7_... 95 2e-17
UniRef100_B3P7W4 GG11910 n=1 Tax=Drosophila erecta RepID=B3P7W4_... 95 2e-17
UniRef100_A1CWE4 Serine/threonine protein kinase, putative n=1 T... 95 2e-17
UniRef100_B6QQQ3 Serine/threonine protein kinase, putative n=1 T... 94 2e-17
UniRef100_B6HTY2 Pc22g04990 protein n=1 Tax=Penicillium chrysoge... 94 2e-17
UniRef100_UPI00016E20E1 UPI00016E20E1 related cluster n=1 Tax=Ta... 94 3e-17
UniRef100_UPI00016E20C4 UPI00016E20C4 related cluster n=1 Tax=Ta... 94 3e-17
UniRef100_UPI00016E20C3 UPI00016E20C3 related cluster n=1 Tax=Ta... 94 3e-17
UniRef100_UPI00016E20C2 UPI00016E20C2 related cluster n=1 Tax=Ta... 94 3e-17
UniRef100_B4LW76 GJ22984 n=1 Tax=Drosophila virilis RepID=B4LW76... 94 3e-17
UniRef100_Q5RIL6 LATS, large tumor suppressor, homolog 1 (Drosop... 94 4e-17
UniRef100_Q58FB8 Large tumor suppressor n=1 Tax=Danio rerio RepI... 94 4e-17
UniRef100_Q0CR08 Serine/threonine-protein kinase cot-1 n=1 Tax=A... 94 4e-17
UniRef100_C1G4B8 Serine/threonine-protein kinase cot-1 n=1 Tax=P... 94 4e-17
UniRef100_C0S595 Serine/threonine-protein kinase cot-1 n=1 Tax=P... 94 4e-17
UniRef100_UPI00006CBD47 Protein kinase domain containing protein... 93 5e-17
UniRef100_Q21613 Ndr protein kinase (Fragment) n=1 Tax=Caenorhab... 93 5e-17
UniRef100_C9SN89 Serine/threonine-protein kinase cot-1 n=1 Tax=V... 93 5e-17
UniRef100_UPI0001757DA9 PREDICTED: similar to GA11375-PA n=1 Tax... 93 7e-17
UniRef100_UPI0000DB6FB9 PREDICTED: similar to warts CG12072-PA n... 93 7e-17
UniRef100_UPI0001985257 PREDICTED: hypothetical protein n=1 Tax=... 92 9e-17
UniRef100_B3MT80 GF23311 n=1 Tax=Drosophila ananassae RepID=B3MT... 92 9e-17
UniRef100_A6RR04 Putative uncharacterized protein n=1 Tax=Botryo... 92 9e-17
UniRef100_UPI00017931CD PREDICTED: similar to mCG22338 n=1 Tax=A... 92 1e-16
UniRef100_UPI00016E7AD0 UPI00016E7AD0 related cluster n=1 Tax=Ta... 92 1e-16
UniRef100_Q4LB97 Putative extensin (Fragment) n=1 Tax=Nicotiana ... 92 1e-16
UniRef100_C5YWZ0 Putative uncharacterized protein Sb09g030460 n=... 92 1e-16
UniRef100_Q9VA38 Warts n=1 Tax=Drosophila melanogaster RepID=Q9V... 92 1e-16
UniRef100_Q86NM6 SD19495p n=1 Tax=Drosophila melanogaster RepID=... 92 1e-16
UniRef100_Q24590 Tumor suppressor n=1 Tax=Drosophila melanogaste... 92 1e-16
UniRef100_Q24096 LATS n=1 Tax=Drosophila melanogaster RepID=Q240... 92 1e-16
UniRef100_C5LYG6 Protein kinase, putative n=1 Tax=Perkinsus mari... 92 1e-16
UniRef100_B4R2A0 GD16829 n=1 Tax=Drosophila simulans RepID=B4R2A... 92 1e-16
UniRef100_B4HZV7 GM12129 n=1 Tax=Drosophila sechellia RepID=B4HZ... 92 1e-16
UniRef100_A8XZW6 C. briggsae CBR-WTS-1 protein n=1 Tax=Caenorhab... 92 1e-16
UniRef100_C0NJN4 Serine/threonine-protein kinase cot-1 n=1 Tax=A... 92 1e-16
UniRef100_A6QVU4 Serine/threonine-protein kinase cot-1 n=1 Tax=A... 92 1e-16
UniRef100_A1CIB6 Serine/threonine protein kinase, putative n=1 T... 92 1e-16
UniRef100_UPI000186DE84 serine/threonine-protein kinase LATS1, p... 92 2e-16
UniRef100_UPI00015B57D1 PREDICTED: similar to GA11375-PA n=1 Tax... 92 2e-16
UniRef100_UPI0000E48661 PREDICTED: similar to large tumor suppre... 92 2e-16
UniRef100_B4JY27 GH14097 n=1 Tax=Drosophila grimshawi RepID=B4JY... 92 2e-16
UniRef100_B2VSH4 Serine/threonine-protein kinase cot-1 n=1 Tax=P... 92 2e-16
UniRef100_B0Y4F3 Serine/threonine protein kinase, putative n=2 T... 92 2e-16
UniRef100_UPI000194B89A PREDICTED: LATS, large tumor suppressor,... 91 2e-16
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 91 2e-16
UniRef100_Q7PQE9 AGAP003618-PA (Fragment) n=1 Tax=Anopheles gamb... 91 2e-16
UniRef100_B4K9G4 GI10631 n=1 Tax=Drosophila mojavensis RepID=B4K... 91 2e-16
UniRef100_A2EKU4 AGC family protein kinase n=1 Tax=Trichomonas v... 91 2e-16
UniRef100_C5GWR5 Serine/threonine-protein kinase cot-1 n=2 Tax=A... 91 2e-16
UniRef100_UPI0000DD9D0C Os11g0657400 n=1 Tax=Oryza sativa Japoni... 91 3e-16
UniRef100_B9G8N7 Putative uncharacterized protein n=1 Tax=Oryza ... 91 3e-16
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 91 3e-16
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 91 3e-16
UniRef100_Q16I19 Serine/threonine protein kinase lats (Fragment)... 91 3e-16
UniRef100_B4NGZ6 GK13110 n=1 Tax=Drosophila willistoni RepID=B4N... 91 3e-16
UniRef100_B0X6M5 Serine/threonine protein kinase lats (Fragment)... 91 3e-16
UniRef100_UPI0000E7FC05 PREDICTED: similar to LATS, large tumor ... 91 3e-16
UniRef100_UPI0000ECD68E Serine/threonine-protein kinase LATS2 (E... 91 3e-16
UniRef100_UPI0000ECD68D Serine/threonine-protein kinase LATS2 (E... 91 3e-16
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 91 3e-16
UniRef100_B7GAH6 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 91 3e-16
UniRef100_Q2UJY3 NDR and related serine/threonine kinases n=1 Ta... 91 3e-16
UniRef100_Q0ULZ0 Putative uncharacterized protein n=1 Tax=Phaeos... 91 3e-16
UniRef100_C5FUA0 Serine/threonine-protein kinase cot-1 n=1 Tax=M... 91 3e-16
UniRef100_B8N116 Serine/threonine protein kinase, putative n=2 T... 91 3e-16
UniRef100_A7E7B7 Putative uncharacterized protein n=1 Tax=Sclero... 91 3e-16
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 90 4e-16
UniRef100_O45797 Protein T20F10.1, partially confirmed by transc... 90 4e-16
UniRef100_A9UVF9 Predicted protein (Fragment) n=1 Tax=Monosiga b... 90 4e-16
UniRef100_UPI0001797323 PREDICTED: LATS, large tumor suppressor,... 90 6e-16
UniRef100_UPI0000F2CE74 PREDICTED: similar to LATS, large tumor ... 90 6e-16
UniRef100_UPI0000E23528 PREDICTED: LATS, large tumor suppressor,... 90 6e-16
UniRef100_UPI0000D9E786 PREDICTED: similar to LATS, large tumor ... 90 6e-16
UniRef100_UPI0000D9E785 PREDICTED: similar to LATS, large tumor ... 90 6e-16
UniRef100_UPI00004BE32F Serine/threonine-protein kinase LATS2 (E... 90 6e-16
UniRef100_UPI00003699D1 PREDICTED: LATS, large tumor suppressor,... 90 6e-16
UniRef100_UPI00001819D6 large tumor suppressor 2 n=1 Tax=Rattus ... 90 6e-16
UniRef100_Q59EB4 LATS, large tumor suppressor, homolog 2 variant... 90 6e-16
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 90 6e-16
UniRef100_Q7TSJ6 Serine/threonine-protein kinase LATS2 n=1 Tax=M... 90 6e-16
UniRef100_Q9NRM7 Serine/threonine-protein kinase LATS2 n=1 Tax=H... 90 6e-16
UniRef100_Q6CFS5 Serine/threonine-protein kinase CBK1 n=1 Tax=Ya... 90 6e-16
UniRef100_UPI0001A7B3CA lipid binding / structural constituent o... 89 1e-15
UniRef100_UPI0001A7B0BB lipid binding / structural constituent o... 89 1e-15
UniRef100_O23370 Cell wall protein like n=1 Tax=Arabidopsis thal... 89 1e-15
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 89 1e-15
UniRef100_UPI0001927592 PREDICTED: similar to serine/threonine k... 89 1e-15
UniRef100_Q01947 Extensin (Class II) n=1 Tax=Solanum lycopersicu... 89 1e-15
UniRef100_A8BB37 Kinase, AGC NDR n=1 Tax=Giardia lamblia ATCC 50... 89 1e-15
UniRef100_A8N8Y0 Putative uncharacterized protein n=1 Tax=Coprin... 89 1e-15
UniRef100_Q9FLQ7 Formin-like protein 20 n=1 Tax=Arabidopsis thal... 89 1e-15
UniRef100_UPI000018A12A serine/threonine-protein kinase orb6 n=1... 88 2e-15
UniRef100_B9RQU4 Putative uncharacterized protein n=1 Tax=Ricinu... 88 2e-15
UniRef100_A7PES6 Chromosome chr11 scaffold_13, whole genome shot... 88 2e-15
UniRef100_C6LUJ7 Kinase, AGC NDR n=1 Tax=Giardia intestinalis AT... 88 2e-15
UniRef100_P38679-2 Isoform Short of Serine/threonine-protein kin... 88 2e-15
UniRef100_P38679 Serine/threonine-protein kinase cot-1 n=1 Tax=N... 88 2e-15
UniRef100_UPI0000EBD489 PREDICTED: similar to LATS, large tumor ... 88 2e-15
UniRef100_Q9SPM0 Extensin-like protein n=1 Tax=Zea mays RepID=Q9... 88 2e-15
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 87 3e-15
UniRef100_Q5ZB68 Os01g0594300 protein n=1 Tax=Oryza sativa Japon... 87 3e-15
UniRef100_Q7PNI8 AGAP000892-PA (Fragment) n=1 Tax=Anopheles gamb... 87 3e-15
UniRef100_C8VG83 COTA [Source:UniProtKB/TrEMBL;Acc:Q6IVG5] n=3 T... 87 3e-15
UniRef100_Q5K2R7 Putative serine/threonine kinase n=1 Tax=Clavic... 87 3e-15
UniRef100_Q00611 Kinase n=1 Tax=Colletotrichum trifolii RepID=Q0... 87 3e-15
UniRef100_Q43586 PAP8 product (Fragment) n=1 Tax=Nicotiana tabac... 87 4e-15
UniRef100_Q43505 Extensin-like protein Dif54 n=1 Tax=Solanum lyc... 87 4e-15
UniRef100_B9RNJ0 Nutrient reservoir, putative n=1 Tax=Ricinus co... 87 4e-15
UniRef100_UPI00015607BB PREDICTED: LATS, large tumor suppressor,... 87 5e-15
UniRef100_Q4TAU7 Chromosome undetermined SCAF7253, whole genome ... 87 5e-15
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 87 5e-15
UniRef100_B9IKQ3 Predicted protein n=1 Tax=Populus trichocarpa R... 87 5e-15
UniRef100_B8C613 Putative uncharacterized protein (Fragment) n=1... 87 5e-15
UniRef100_Q9VZC0 CG11345 n=1 Tax=Drosophila melanogaster RepID=Q... 87 5e-15
UniRef100_B2AN55 Predicted CDS Pa_6_9030 n=1 Tax=Podospora anser... 87 5e-15
UniRef100_UPI00006A2209 UPI00006A2209 related cluster n=1 Tax=Xe... 86 6e-15
UniRef100_Q641H7 MGC81565 protein n=1 Tax=Xenopus laevis RepID=Q... 86 6e-15
UniRef100_Q8LJ87 Os01g0356900 protein n=1 Tax=Oryza sativa Japon... 86 6e-15
UniRef100_B4J560 GH20878 n=1 Tax=Drosophila grimshawi RepID=B4J5... 86 6e-15
UniRef100_Q2H1B2 Serine/threonine-protein kinase cot-1 n=1 Tax=C... 86 6e-15
UniRef100_A4III9 Lats2 protein n=1 Tax=Xenopus (Silurana) tropic... 86 8e-15