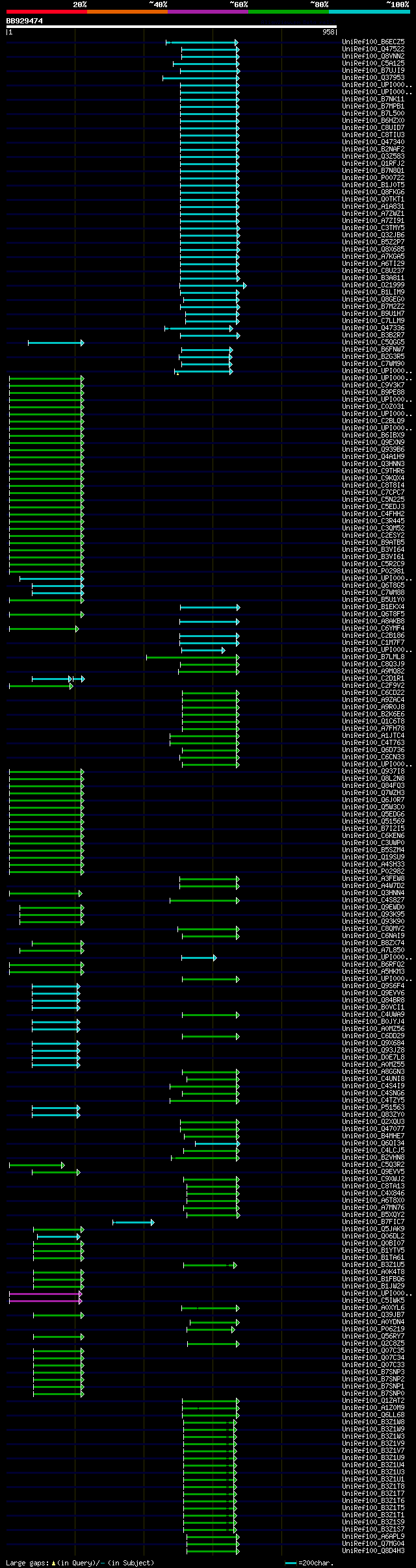
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB929474 RCC00916
(958 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B6ECZ5 LacZ alpha n=1 Tax=Cloning vector pMAK28 RepID=... 117 2e-24
UniRef100_Q47522 LacZ protein (Fragment) n=1 Tax=Escherichia col... 117 2e-24
UniRef100_Q8VNN2 Beta-galactosidase n=1 Tax=Escherichia coli Rep... 117 2e-24
UniRef100_C5A125 Tryptophan synthase alpha chain n=1 Tax=Escheri... 117 2e-24
UniRef100_B7UJI9 Beta-galactosidase n=1 Tax=Escherichia coli O12... 117 2e-24
UniRef100_Q37953 LacZ protein (Fragment) n=1 Tax=Phage M13mp18 R... 116 4e-24
UniRef100_UPI0001B5303F beta-D-galactosidase n=1 Tax=Shigella sp... 115 5e-24
UniRef100_UPI0001B525AD beta-D-galactosidase n=1 Tax=Escherichia... 115 5e-24
UniRef100_B7NK11 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 115 5e-24
UniRef100_B7MPB1 Beta-D-galactosidase n=1 Tax=Escherichia coli E... 115 5e-24
UniRef100_B7L500 Beta-D-galactosidase n=1 Tax=Escherichia coli 5... 115 5e-24
UniRef100_B6HZX0 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 115 5e-24
UniRef100_C8UID7 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 115 5e-24
UniRef100_C8TIU3 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 115 5e-24
UniRef100_Q47340 LacZ 5'-region (Fragment) n=2 Tax=Escherichia c... 115 5e-24
UniRef100_B2NAF2 Beta-galactosidase n=1 Tax=Escherichia coli 536... 115 5e-24
UniRef100_Q3Z583 Beta-galactosidase n=1 Tax=Shigella sonnei Ss04... 115 5e-24
UniRef100_Q1RFJ2 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 115 5e-24
UniRef100_B7N8Q1 Beta-galactosidase n=1 Tax=Escherichia coli UMN... 115 5e-24
UniRef100_P00722 Beta-galactosidase n=6 Tax=Escherichia coli Rep... 115 5e-24
UniRef100_B1J0T5 Beta-galactosidase n=1 Tax=Escherichia coli ATC... 115 5e-24
UniRef100_Q8FKG6 Beta-galactosidase n=2 Tax=Escherichia coli Rep... 115 5e-24
UniRef100_Q0TKT1 Beta-galactosidase n=1 Tax=Escherichia coli 536... 115 5e-24
UniRef100_A1A831 Beta-galactosidase n=1 Tax=Escherichia coli APE... 115 5e-24
UniRef100_A7ZWZ1 Beta-galactosidase n=1 Tax=Escherichia coli HS ... 115 5e-24
UniRef100_A7ZI91 Beta-galactosidase n=1 Tax=Escherichia coli E24... 115 5e-24
UniRef100_C3TMY5 Beta-D-galactosidase n=1 Tax=Escherichia coli R... 115 9e-24
UniRef100_Q32JB6 Beta-galactosidase n=1 Tax=Shigella dysenteriae... 115 9e-24
UniRef100_B5Z2P7 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 115 9e-24
UniRef100_Q8X685 Beta-galactosidase n=3 Tax=Escherichia coli Rep... 115 9e-24
UniRef100_A7KGA5 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 114 1e-23
UniRef100_A6TI29 Beta-galactosidase 2 n=1 Tax=Klebsiella pneumon... 114 1e-23
UniRef100_C8U237 Beta-D-galactosidase LacZ n=1 Tax=Escherichia c... 114 2e-23
UniRef100_B3A811 Beta-galactosidase n=1 Tax=Escherichia coli O15... 114 2e-23
UniRef100_O21999 LacZ-alpha n=1 Tax=Cloning vector pAL-Z RepID=O... 113 3e-23
UniRef100_B1LIM9 Beta-galactosidase n=1 Tax=Escherichia coli SMS... 112 6e-23
UniRef100_Q8GEG0 Putative uncharacterized protein n=1 Tax=Erwini... 111 1e-22
UniRef100_B7M2Z2 Beta-D-galactosidase n=1 Tax=Escherichia coli I... 111 1e-22
UniRef100_B9U1H7 Beta-D-galactosidase n=1 Tax=Vaccinia virus GLV... 110 2e-22
UniRef100_C7LLM9 Beta-galactosidase, chain D n=1 Tax=Mycoplasma ... 110 2e-22
UniRef100_Q47336 LacZ-alpha peptide n=1 Tax=Escherichia coli Rep... 106 4e-21
UniRef100_B3B2R7 Beta-galactosidase n=1 Tax=Escherichia coli O15... 106 4e-21
UniRef100_C5QGG5 MFS family major facilitator tetracyline transp... 104 1e-20
UniRef100_B6FNW7 Putative uncharacterized protein n=1 Tax=Clostr... 103 3e-20
UniRef100_B2G3R5 Putative puroindoline b protein n=1 Tax=Triticu... 103 4e-20
UniRef100_C7WM90 LacZ alpha protein (Fragment) n=1 Tax=Enterococ... 102 8e-20
UniRef100_UPI0000ECC20B UPI0000ECC20B related cluster n=1 Tax=Ga... 101 1e-19
UniRef100_UPI0001B49BF3 tetracycline resistance protein n=1 Tax=... 100 3e-19
UniRef100_C9V3K7 Tetracycline resistance protein (Fragment) n=2 ... 100 3e-19
UniRef100_B9PE88 Predicted protein n=3 Tax=cellular organisms Re... 100 3e-19
UniRef100_UPI0001AF5C09 tetracycline resistance protein n=1 Tax=... 100 3e-19
UniRef100_C0Z031 MFS family major facilitator tetracyline transp... 100 3e-19
UniRef100_UPI0001AF437D TetA n=1 Tax=Neisseria gonorrhoeae PID24... 100 3e-19
UniRef100_C2BLQ9 MFS family major facilitator tetracyline transp... 100 3e-19
UniRef100_UPI0001660031 TetA n=1 Tax=Escherichia coli RepID=UPI0... 100 3e-19
UniRef100_B6IBX9 Tetracycline resistance structural protein TetA... 100 3e-19
UniRef100_Q9EXN9 Tetracycline resistance (Fragment) n=1 Tax=Esch... 100 3e-19
UniRef100_Q939B6 Tetracycline resistance structural protein TetA... 100 3e-19
UniRef100_Q4A1H9 Tetracycline resistance protein n=2 Tax=Bacteri... 100 3e-19
UniRef100_Q3HNN3 Tetracycline resistance protein (Fragment) n=4 ... 100 3e-19
UniRef100_C9THR6 Tetracycline resistance protein n=1 Tax=Brucell... 100 3e-19
UniRef100_C9KQX4 Tetracycline-efflux transporter (Fragment) n=1 ... 100 3e-19
UniRef100_C8T8I4 Tetracycline resistance protein (Fragment) n=1 ... 100 3e-19
UniRef100_C7CPC7 Tetracycline resistance protein n=5 Tax=Bacteri... 100 3e-19
UniRef100_C5N225 Possible tetracycline resistance structural pro... 100 3e-19
UniRef100_C5EDJ3 Tetracycline resistance protein (Fragment) n=2 ... 100 3e-19
UniRef100_C4FHH2 Putative uncharacterized protein (Fragment) n=1... 100 3e-19
UniRef100_C3R445 Tetracycline resistance protein (Fragment) n=2 ... 100 3e-19
UniRef100_C3QM52 Tetracycline resistance protein n=2 Tax=Bacteri... 100 3e-19
UniRef100_C2ESY2 MFS family major facilitator tetracyline transp... 100 3e-19
UniRef100_B9ATB5 Putative uncharacterized protein (Fragment) n=1... 100 3e-19
UniRef100_B3VI64 Tetracycline resistance protein n=2 Tax=root Re... 100 3e-19
UniRef100_B3VI61 Tetracycline resistance protein n=22 Tax=root R... 100 3e-19
UniRef100_C5R2C9 MFS family major facilitator tetracyline transp... 100 3e-19
UniRef100_P02981 Tetracycline resistance protein, class C n=3 Ta... 100 3e-19
UniRef100_UPI0001B48888 tetracycline resistance protein n=1 Tax=... 99 5e-19
UniRef100_Q6T8G5 Tet(C) n=1 Tax=Chlamydia suis RepID=Q6T8G5_9CHLA 99 7e-19
UniRef100_C7WM88 Tetracycline efflux protein (Fragment) n=1 Tax=... 99 7e-19
UniRef100_B5U1Y0 Tetracycline resistance protein n=1 Tax=uncultu... 98 1e-18
UniRef100_B1EKX4 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 97 2e-18
UniRef100_Q6T8F5 Tet(C) n=1 Tax=Chlamydia suis RepID=Q6T8F5_9CHLA 96 7e-18
UniRef100_A8AKB8 Beta-galactosidase n=1 Tax=Citrobacter koseri A... 94 2e-17
UniRef100_C6YMF4 Tetracycline resistance protein (Fragment) n=2 ... 94 3e-17
UniRef100_C2B186 Putative uncharacterized protein n=1 Tax=Citrob... 92 8e-17
UniRef100_C1M7F7 Beta-D-galactosidase n=1 Tax=Citrobacter sp. 30... 92 8e-17
UniRef100_UPI0000ECBAF8 UPI0000ECBAF8 related cluster n=1 Tax=Ga... 89 5e-16
UniRef100_B7LML8 Beta-galactosidase (Lactase) n=1 Tax=Escherichi... 87 3e-15
UniRef100_C8Q3J9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 86 8e-15
UniRef100_A9MQ82 Beta-galactosidase n=1 Tax=Salmonella enterica ... 85 1e-14
UniRef100_C2D1R1 Putative uncharacterized protein (Fragment) n=1... 80 2e-14
UniRef100_C2F9V2 Putative uncharacterized protein (Fragment) n=1... 83 4e-14
UniRef100_C6CD22 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 81 2e-13
UniRef100_A9ZAC4 Beta-galactosidase (Lactase) n=2 Tax=Yersinia p... 78 1e-12
UniRef100_A9R0J8 Beta-galactosidase n=6 Tax=Yersinia pestis RepI... 78 1e-12
UniRef100_B2K6E6 Beta-galactosidase n=3 Tax=Yersinia pseudotuber... 78 1e-12
UniRef100_Q1C6T8 Beta-galactosidase n=8 Tax=Yersinia pestis RepI... 78 1e-12
UniRef100_A7FH78 Beta-galactosidase n=1 Tax=Yersinia pseudotuber... 78 1e-12
UniRef100_A1JTC4 Beta-galactosidase n=1 Tax=Yersinia enterocolit... 78 1e-12
UniRef100_C4T763 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 78 2e-12
UniRef100_Q6D736 Beta-galactosidase n=1 Tax=Pectobacterium atros... 77 3e-12
UniRef100_C6CN33 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 77 3e-12
UniRef100_UPI0001826C8D hypothetical protein ENTCAN_01162 n=1 Ta... 76 5e-12
UniRef100_Q937I8 Tetracycline efflux protein n=1 Tax=Escherichia... 76 6e-12
UniRef100_Q8L2N8 Tetracycline resistance protein A n=1 Tax=Shige... 76 6e-12
UniRef100_Q84FQ3 Tetracycline resistance protein A (Fragment) n=... 76 6e-12
UniRef100_Q7WZH3 Tetracycline resistance protein A n=1 Tax=Vibri... 76 6e-12
UniRef100_Q6J0R7 TetA n=1 Tax=Escherichia coli RepID=Q6J0R7_ECOLX 76 6e-12
UniRef100_Q5W3C0 Tetracycline efflux protein n=1 Tax=uncultured ... 76 6e-12
UniRef100_Q5EDG6 Tetracycline resistance protein A n=1 Tax=Larib... 76 6e-12
UniRef100_Q51569 Tetracycline exporter protein n=3 Tax=root RepI... 76 6e-12
UniRef100_B7I2I5 Tetracycline resistance protein, class A n=3 Ta... 76 6e-12
UniRef100_C6KEN6 Tetracycline resistance protein class A (Fragme... 76 6e-12
UniRef100_C3UWP0 Tetracycline resistance protein class A (Fragme... 76 6e-12
UniRef100_B5SZM4 Tetracycline resistance efflux pump n=1 Tax=Sal... 76 6e-12
UniRef100_Q19SU9 Tetracycline efflux protein TetA n=16 Tax=root ... 76 6e-12
UniRef100_A4SH33 TetA n=5 Tax=Enterobacteriaceae RepID=A4SH33_SHISS 76 6e-12
UniRef100_P02982 Tetracycline resistance protein, class A n=17 T... 76 6e-12
UniRef100_A3FEW8 Beta-galactosidase n=1 Tax=Pantoea agglomerans ... 76 6e-12
UniRef100_A4W7D2 Beta-galactosidase n=1 Tax=Enterobacter sp. 638... 76 6e-12
UniRef100_Q3HNN4 Tetracycline resistance protein (Fragment) n=1 ... 75 8e-12
UniRef100_C4S827 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 75 8e-12
UniRef100_Q9EWD0 Tetracycline resistance (Fragment) n=1 Tax=Salm... 75 1e-11
UniRef100_Q93K95 Tetracycline resistance protein of class A n=1 ... 75 1e-11
UniRef100_Q93K90 Tetracycline resistance protein, class A n=1 Ta... 75 1e-11
UniRef100_C8QMV2 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 75 1e-11
UniRef100_C6NAI9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 75 1e-11
UniRef100_B8ZX74 Tetracycline resistance protein Tet(A) (Fragmen... 75 1e-11
UniRef100_A7L850 TetA n=1 Tax=Klebsiella pneumoniae RepID=A7L850... 75 1e-11
UniRef100_UPI0000ECAFAB UPI0000ECAFAB related cluster n=1 Tax=Ga... 74 2e-11
UniRef100_B6RFQ2 Tetracycline resistance protein n=1 Tax=Klebsie... 74 3e-11
UniRef100_A5HKM3 TetA (Fragment) n=1 Tax=Salmonella enterica sub... 74 3e-11
UniRef100_UPI0001A43550 beta-D-galactosidase n=1 Tax=Pectobacter... 73 4e-11
UniRef100_Q9S6F4 Tetracycline resistance protein TetG (Fragment)... 73 4e-11
UniRef100_Q9EVV6 Tetracycline resistance (Fragment) n=1 Tax=Salm... 73 4e-11
UniRef100_Q84BR8 TetG protein n=1 Tax=Stenotrophomonas sp. TA57 ... 73 4e-11
UniRef100_B0VCI1 Tetracycline resistance protein, class G (TETA(... 73 4e-11
UniRef100_C4UWA9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 73 4e-11
UniRef100_B0JYJ4 TetA(G) n=3 Tax=Salmonella enterica subsp. ente... 73 4e-11
UniRef100_A0MZ56 Tetracycline resistance protein (Fragment) n=1 ... 73 4e-11
UniRef100_C6DD29 Beta-galactosidase n=1 Tax=Pectobacterium carot... 73 5e-11
UniRef100_Q9X684 Tetracycline resistance protein n=1 Tax=Pseudom... 73 5e-11
UniRef100_Q93JZ8 Tetracycline resistance protein, class G (Fragm... 73 5e-11
UniRef100_D0E7L8 Tetracycline efflux pump n=1 Tax=Pseudomonas ae... 73 5e-11
UniRef100_A0MZ55 Tetracycline resistance protein (Fragment) n=1 ... 73 5e-11
UniRef100_A8GGN3 Beta-galactosidase n=1 Tax=Serratia proteamacul... 73 5e-11
UniRef100_C4UNI8 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 7e-11
UniRef100_C4S4I9 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 7e-11
UniRef100_C4SNG6 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 9e-11
UniRef100_C4TZY5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 72 1e-10
UniRef100_P51563 Tetracycline resistance protein, class G n=1 Ta... 72 1e-10
UniRef100_Q83ZY0 Tetracycline resistance protein n=1 Tax=Pasteur... 71 1e-10
UniRef100_Q2XQU3 Beta-galactosidase 2 n=1 Tax=Enterobacter cloac... 71 1e-10
UniRef100_Q47077 Beta-galactosidase n=1 Tax=Enterobacter cloacae... 71 1e-10
UniRef100_B4MHE7 GJ14580 n=1 Tax=Drosophila virilis RepID=B4MHE7... 71 1e-10
UniRef100_Q6QI34 LRRGT00174 n=1 Tax=Rattus norvegicus RepID=Q6QI... 71 2e-10
UniRef100_C4LCJ5 Glycoside hydrolase family 2 TIM barrel n=1 Tax... 70 3e-10
UniRef100_B2VHN8 Beta-galactosidase n=1 Tax=Erwinia tasmaniensis... 69 7e-10
UniRef100_C5Q3R2 Putative uncharacterized protein (Fragment) n=1... 67 2e-09
UniRef100_Q9EVV5 Tetracycline resistance (Fragment) n=1 Tax=Salm... 67 4e-09
UniRef100_C9XWJ2 Beta-galactosidase n=1 Tax=Cronobacter turicens... 67 4e-09
UniRef100_C8TA13 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 67 4e-09
UniRef100_C4X846 Beta-D-galactosidase n=1 Tax=Klebsiella pneumon... 67 4e-09
UniRef100_A6T8X0 Beta-galactosidase 1 n=1 Tax=Klebsiella pneumon... 67 4e-09
UniRef100_A7MN76 Beta-galactosidase n=1 Tax=Cronobacter sakazaki... 66 6e-09
UniRef100_B5XQY2 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 65 1e-08
UniRef100_B7FIC7 Putative uncharacterized protein n=1 Tax=Medica... 62 7e-08
UniRef100_Q5JAK9 TetA(41) n=1 Tax=Serratia marcescens RepID=Q5JA... 60 3e-07
UniRef100_Q06DL2 TetG (Fragment) n=1 Tax=Brevundimonas sp. PB-P1... 60 3e-07
UniRef100_Q0BI07 Major facilitator superfamily MFS_1 n=1 Tax=Bur... 59 6e-07
UniRef100_B1YTV5 Major facilitator superfamily MFS_1 n=1 Tax=Bur... 59 6e-07
UniRef100_B1TA61 Major facilitator superfamily MFS_1 n=1 Tax=Bur... 59 6e-07
UniRef100_B3Z1U5 Putative uncharacterized protein n=1 Tax=Bacill... 59 1e-06
UniRef100_A0K4T8 Major facilitator superfamily MFS_1 n=2 Tax=Bur... 58 1e-06
UniRef100_B1FBQ6 Major facilitator superfamily MFS_1 n=1 Tax=Bur... 58 1e-06
UniRef100_B1JW29 Major facilitator superfamily MFS_1 n=2 Tax=Bur... 58 1e-06
UniRef100_UPI000197CCB9 hypothetical protein PROVRETT_04001 n=1 ... 58 2e-06
UniRef100_C5IWK5 Tetracycline resistance protein n=3 Tax=Gammapr... 58 2e-06
UniRef100_A0XYL6 Beta-D-galactosidase n=1 Tax=Alteromonadales ba... 57 2e-06
UniRef100_Q39JB7 Major facilitator superfamily (MFS_1) transport... 57 3e-06
UniRef100_A0YDN4 Beta-D-galactosidase n=1 Tax=marine gamma prote... 57 3e-06
UniRef100_P06219 Beta-galactosidase n=1 Tax=Klebsiella pneumonia... 57 3e-06
UniRef100_Q56RY7 TetA(39) n=1 Tax=Acinetobacter sp. LUH5605 RepI... 57 4e-06
UniRef100_Q2C8Z5 Beta-galactosidase n=1 Tax=Photobacterium sp. S... 57 4e-06
UniRef100_Q07C35 Tetracycline resistance efflux pump (Fragment) ... 57 4e-06
UniRef100_Q07C34 Tetracycline resistance efflux pump (Fragment) ... 57 4e-06
UniRef100_Q07C33 Tetracycline resistance efflux pump (Fragment) ... 57 4e-06
UniRef100_B7SNP3 Tetracycline efflux protein (Fragment) n=5 Tax=... 57 4e-06
UniRef100_B7SNP2 Tetracycline efflux protein (Fragment) n=1 Tax=... 57 4e-06
UniRef100_B7SNP1 Tetracycline efflux protein (Fragment) n=1 Tax=... 57 4e-06
UniRef100_B7SNP0 Tetracycline efflux protein (Fragment) n=1 Tax=... 57 4e-06
UniRef100_Q1ZAT2 Putative beta-galactosidase n=1 Tax=Photobacter... 56 5e-06
UniRef100_A1Z0M9 Beta-galactosidase n=1 Tax=Pseudoalteromonas sp... 56 5e-06
UniRef100_Q6LL68 Beta-galactosidase n=1 Tax=Photobacterium profu... 56 5e-06
UniRef100_B3Z1W8 Putative uncharacterized protein (Fragment) n=1... 56 5e-06
UniRef100_B3Z1W9 Putative uncharacterized protein (Fragment) n=1... 56 6e-06
UniRef100_B3Z1W3 Putative uncharacterized protein (Fragment) n=1... 56 6e-06
UniRef100_B3Z1V9 Putative uncharacterized protein (Fragment) n=1... 56 6e-06
UniRef100_B3Z1V7 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1U9 Ferrichrome-binding protein (Fragment) n=1 Tax=... 56 6e-06
UniRef100_B3Z1U4 Putative uncharacterized protein (Fragment) n=1... 56 6e-06
UniRef100_B3Z1U3 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1U1 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1T8 Endopeptidase lactocepin n=1 Tax=Bacillus cereu... 56 6e-06
UniRef100_B3Z1T7 Cell-division initiation protein DivIB n=1 Tax=... 56 6e-06
UniRef100_B3Z1T6 Amino acid permease n=1 Tax=Bacillus cereus W R... 56 6e-06
UniRef100_B3Z1T5 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1T1 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1S9 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_B3Z1S7 Putative uncharacterized protein n=1 Tax=Bacill... 56 6e-06
UniRef100_A6APL9 Beta-galactosidase n=1 Tax=Vibrio harveyi HY01 ... 55 8e-06
UniRef100_Q7MG04 Beta-galactosidase n=1 Tax=Vibrio vulnificus YJ... 55 8e-06
UniRef100_Q8D4H3 Beta-galactosidase n=1 Tax=Vibrio vulnificus Re... 55 8e-06
UniRef100_P81650 Beta-galactosidase n=1 Tax=Pseudoalteromonas ha... 55 8e-06