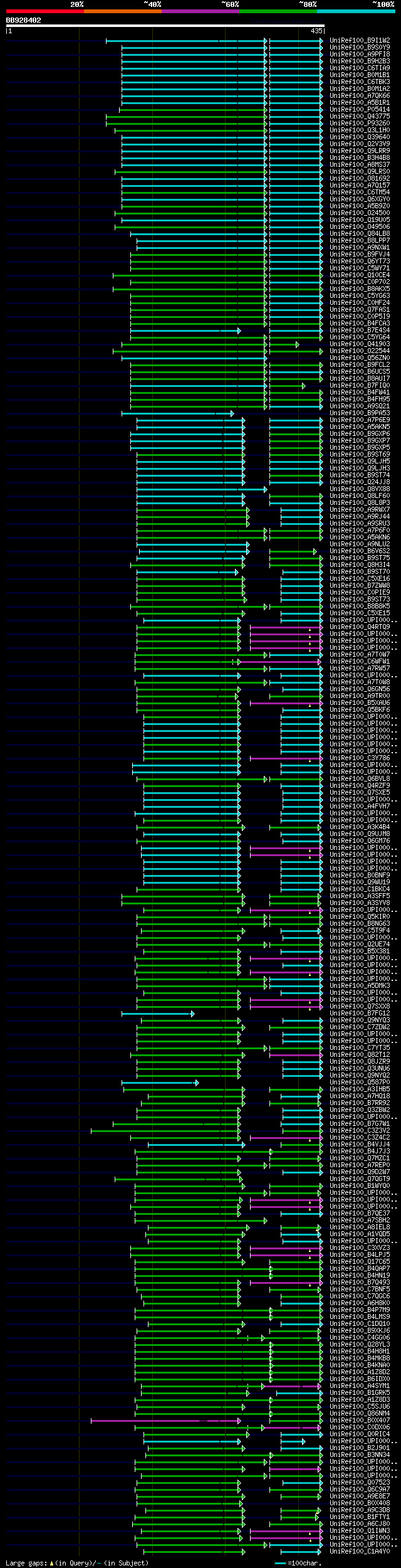
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB928402 RCE39604
(435 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9I1W2 Predicted protein n=3 Tax=Populus RepID=B9I1W2_... 102 5e-27
UniRef100_B9S0Y9 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 96 3e-25
UniRef100_A9PFI8 Predicted protein n=1 Tax=Populus trichocarpa R... 96 3e-25
UniRef100_B9H2B3 Predicted protein n=1 Tax=Populus trichocarpa R... 94 4e-25
UniRef100_C6TIA9 Putative uncharacterized protein n=1 Tax=Glycin... 97 4e-25
UniRef100_B0M1B1 Peroxisomal glycolate oxidase n=1 Tax=Glycine m... 97 6e-25
UniRef100_C6TBK3 Putative uncharacterized protein n=1 Tax=Glycin... 97 7e-25
UniRef100_B0M1A2 Peroxisomal glycolate oxidase n=1 Tax=Glycine m... 97 7e-25
UniRef100_A7QK66 Chromosome chr19 scaffold_111, whole genome sho... 94 1e-24
UniRef100_A5B1R1 Putative uncharacterized protein n=1 Tax=Vitis ... 94 1e-24
UniRef100_P05414 Peroxisomal (S)-2-hydroxy-acid oxidase n=1 Tax=... 94 1e-24
UniRef100_Q43775 Glycolate oxidase (Fragment) n=1 Tax=Solanum ly... 96 1e-24
UniRef100_P93260 Glycolate oxidase n=1 Tax=Mesembryanthemum crys... 93 2e-24
UniRef100_Q3L1H0 Glycolate oxidase n=1 Tax=Brassica napus RepID=... 93 3e-24
UniRef100_Q39640 Glycolate oxidase n=1 Tax=Cucurbita cv. Kurokaw... 96 5e-24
UniRef100_Q2V3V9 Uncharacterized protein At3g14420.3 n=1 Tax=Ara... 92 6e-24
UniRef100_Q9LRR9 Probable peroxisomal (S)-2-hydroxy-acid oxidase... 92 6e-24
UniRef100_B3H4B8 Uncharacterized protein At3g14420.6 n=1 Tax=Ara... 92 6e-24
UniRef100_A8MS37 Uncharacterized protein At3g14420.5 n=1 Tax=Ara... 92 6e-24
UniRef100_Q9LRS0 Probable peroxisomal (S)-2-hydroxy-acid oxidase... 92 8e-24
UniRef100_O81692 Glycolate oxidase (Fragment) n=1 Tax=Medicago s... 94 8e-24
UniRef100_A7Q157 Chromosome chr10 scaffold_43, whole genome shot... 90 2e-23
UniRef100_C6TM54 Putative uncharacterized protein n=1 Tax=Glycin... 94 2e-23
UniRef100_Q6XGY0 Putative glycolate oxidase (Fragment) n=1 Tax=V... 90 2e-23
UniRef100_A5B9Z0 Putative uncharacterized protein n=1 Tax=Vitis ... 89 3e-23
UniRef100_O24500 Glycolate oxidase (Fragment) n=1 Tax=Arabidopsi... 91 1e-22
UniRef100_Q19U05 Glycolate oxidase (Fragment) n=1 Tax=Pachysandr... 89 1e-22
UniRef100_O49506 Glycolate oxidase - like protein n=1 Tax=Arabid... 85 5e-22
UniRef100_Q84LB8 Glycolate oxidase n=1 Tax=Zantedeschia aethiopi... 82 4e-21
UniRef100_B8LPP7 Putative uncharacterized protein n=1 Tax=Picea ... 79 4e-20
UniRef100_A9NXW1 Putative uncharacterized protein n=1 Tax=Picea ... 79 4e-20
UniRef100_B9FVJ4 Putative uncharacterized protein n=1 Tax=Oryza ... 82 5e-20
UniRef100_Q6YT73 Os07g0152900 protein n=2 Tax=Oryza sativa RepID... 82 5e-20
UniRef100_C5WY71 Putative uncharacterized protein Sb01g005960 n=... 80 5e-20
UniRef100_Q10CE4 Os03g0786100 protein n=2 Tax=Oryza sativa RepID... 82 3e-19
UniRef100_C0P702 Putative uncharacterized protein n=2 Tax=Zea ma... 81 3e-19
UniRef100_B8AKX5 Putative uncharacterized protein n=1 Tax=Oryza ... 82 3e-19
UniRef100_C5YG63 Putative uncharacterized protein Sb06g028990 n=... 79 2e-18
UniRef100_C0HF24 Putative uncharacterized protein n=1 Tax=Zea ma... 79 2e-18
UniRef100_Q7FAS1 Os04g0623500 protein n=3 Tax=Oryza sativa RepID... 79 2e-18
UniRef100_C0P5I9 Putative uncharacterized protein n=1 Tax=Zea ma... 79 2e-18
UniRef100_B4FCA3 Putative uncharacterized protein n=1 Tax=Zea ma... 79 5e-18
UniRef100_B7E4S4 cDNA clone:001-002-F07, full insert sequence n=... 77 7e-18
UniRef100_C5YG64 Putative uncharacterized protein Sb06g029000 n=... 78 1e-17
UniRef100_Q41903 (S)-2-hydroxy-acid oxidase (Fragment) n=1 Tax=A... 89 1e-17
UniRef100_O22544 Glycolate oxidase n=1 Tax=Oryza sativa RepID=O2... 76 2e-17
UniRef100_Q56ZN0 Glycolate oxidase like protein (Fragment) n=1 T... 92 2e-17
UniRef100_B9FCL2 Putative uncharacterized protein n=1 Tax=Oryza ... 79 6e-17
UniRef100_B6UCS5 Hydroxyacid oxidase 1 n=1 Tax=Zea mays RepID=B6... 75 2e-16
UniRef100_B8AUI7 Putative uncharacterized protein n=1 Tax=Oryza ... 79 2e-16
UniRef100_B7FIQ0 Putative uncharacterized protein n=1 Tax=Medica... 81 4e-16
UniRef100_B4FW41 Putative uncharacterized protein n=1 Tax=Zea ma... 75 5e-16
UniRef100_B4FH95 Putative uncharacterized protein n=1 Tax=Zea ma... 75 5e-16
UniRef100_A9SQ21 Predicted protein n=1 Tax=Physcomitrella patens... 66 2e-15
UniRef100_B9PA53 Predicted protein (Fragment) n=1 Tax=Populus tr... 85 3e-15
UniRef100_A7P6E9 Chromosome chr9 scaffold_7, whole genome shotgu... 67 4e-15
UniRef100_A5AKN5 Putative uncharacterized protein n=1 Tax=Vitis ... 67 4e-15
UniRef100_B9GXP6 Predicted protein n=1 Tax=Populus trichocarpa R... 69 1e-14
UniRef100_B9GXP7 Predicted protein n=1 Tax=Populus trichocarpa R... 69 1e-14
UniRef100_B9GXP5 Predicted protein n=1 Tax=Populus trichocarpa R... 69 1e-14
UniRef100_B9ST69 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 67 1e-14
UniRef100_Q9LJH5 Glycolate oxidase n=1 Tax=Arabidopsis thaliana ... 64 2e-14
UniRef100_Q9LJH3 Glycolate oxidase n=1 Tax=Arabidopsis thaliana ... 64 3e-14
UniRef100_B9ST74 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 69 3e-14
UniRef100_Q24JJ8 At3g14150 n=1 Tax=Arabidopsis thaliana RepID=Q2... 64 3e-14
UniRef100_Q8VX88 Putative (S)-2-hydroxy-acid oxidase (Fragment) ... 81 4e-14
UniRef100_Q8LF60 Glycolate oxidase, putative n=1 Tax=Arabidopsis... 64 5e-14
UniRef100_Q8L8P3 Glycolate oxidase, putative n=1 Tax=Arabidopsis... 64 5e-14
UniRef100_A9RWX7 Predicted protein n=1 Tax=Physcomitrella patens... 62 2e-13
UniRef100_A9RJ44 Predicted protein n=1 Tax=Physcomitrella patens... 62 2e-13
UniRef100_A9SRU3 Predicted protein n=1 Tax=Physcomitrella patens... 61 2e-13
UniRef100_A7P6F0 Chromosome chr9 scaffold_7, whole genome shotgu... 65 2e-13
UniRef100_A5AKN6 Putative uncharacterized protein n=1 Tax=Vitis ... 65 2e-13
UniRef100_A9NLU2 Putative uncharacterized protein n=1 Tax=Picea ... 78 3e-13
UniRef100_B6V6S2 Putative glycolate oxidase (Fragment) n=1 Tax=C... 66 4e-13
UniRef100_B9ST75 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 65 5e-13
UniRef100_Q8H3I4 Os07g0616500 protein n=1 Tax=Oryza sativa Japon... 63 7e-13
UniRef100_B9ST70 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 64 9e-13
UniRef100_C5XE16 Putative uncharacterized protein Sb02g039250 n=... 61 2e-12
UniRef100_B7ZWW8 Putative uncharacterized protein n=1 Tax=Zea ma... 61 2e-12
UniRef100_C0PIE9 Putative uncharacterized protein n=1 Tax=Zea ma... 61 2e-12
UniRef100_B9ST73 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ri... 65 3e-12
UniRef100_B8B8K5 Putative uncharacterized protein n=1 Tax=Oryza ... 60 3e-12
UniRef100_C5XE15 Putative uncharacterized protein Sb02g039240 n=... 60 4e-12
UniRef100_UPI000155D102 PREDICTED: hypothetical protein n=1 Tax=... 55 1e-10
UniRef100_Q4RTQ9 Chromosome 2 SCAF14997, whole genome shotgun se... 56 2e-10
UniRef100_UPI00016E7AFA UPI00016E7AFA related cluster n=1 Tax=Ta... 56 2e-10
UniRef100_UPI00017B3E7C UPI00017B3E7C related cluster n=1 Tax=Te... 56 2e-10
UniRef100_UPI00016E7AFB UPI00016E7AFB related cluster n=1 Tax=Ta... 56 2e-10
UniRef100_A7T0W7 Predicted protein n=1 Tax=Nematostella vectensi... 54 2e-10
UniRef100_C6WFW1 FMN-dependent alpha-hydroxy acid dehydrogenase ... 55 3e-10
UniRef100_A7RW57 Predicted protein n=1 Tax=Nematostella vectensi... 54 5e-10
UniRef100_UPI0000F2B908 PREDICTED: similar to glycolate oxidase;... 54 5e-10
UniRef100_A7T0W8 Predicted protein (Fragment) n=1 Tax=Nematostel... 54 5e-10
UniRef100_Q6GN56 LOC398510 protein n=1 Tax=Xenopus laevis RepID=... 54 8e-10
UniRef100_A9TR00 Predicted protein n=1 Tax=Physcomitrella patens... 54 8e-10
UniRef100_B5XAU6 Hydroxyacid oxidase 2 n=1 Tax=Salmo salar RepID... 53 1e-09
UniRef100_Q5BKF6 MGC108441 protein n=1 Tax=Xenopus (Silurana) tr... 54 1e-09
UniRef100_UPI0000E80025 PREDICTED: hypothetical protein n=1 Tax=... 52 2e-09
UniRef100_UPI0000EB0E34 Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAO... 52 2e-09
UniRef100_UPI00004BE03F PREDICTED: similar to Hydroxyacid oxidas... 52 2e-09
UniRef100_UPI000155FFD5 PREDICTED: hydroxyacid oxidase (glycolat... 52 2e-09
UniRef100_UPI0000ECC94A Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAO... 52 2e-09
UniRef100_UPI00005A4408 PREDICTED: similar to hydroxyacid oxidas... 52 2e-09
UniRef100_C3Y786 Putative uncharacterized protein n=1 Tax=Branch... 52 2e-09
UniRef100_UPI000179DF08 UPI000179DF08 related cluster n=1 Tax=Bo... 52 2e-09
UniRef100_UPI000179DF09 UPI000179DF09 related cluster n=1 Tax=Bo... 52 2e-09
UniRef100_Q6BVL8 DEHA2C01584p n=1 Tax=Debaryomyces hansenii RepI... 49 2e-09
UniRef100_Q4RZF9 Chromosome 3 SCAF14932, whole genome shotgun se... 49 2e-09
UniRef100_Q7SXE5 Hao1 protein (Fragment) n=1 Tax=Danio rerio Rep... 55 2e-09
UniRef100_UPI0000F21F17 hydroxyacid oxidase (glycolate oxidase) ... 55 2e-09
UniRef100_A4FVH7 Hao1 protein n=1 Tax=Danio rerio RepID=A4FVH7_D... 55 2e-09
UniRef100_UPI000179DEF5 UPI000179DEF5 related cluster n=1 Tax=Bo... 51 2e-09
UniRef100_UPI00016E3DF9 UPI00016E3DF9 related cluster n=1 Tax=Ta... 51 3e-09
UniRef100_A3K4B4 Glycolate oxidase, (S)-2-hydroxy-acid oxidase, ... 56 3e-09
UniRef100_Q9UJM8 Hydroxyacid oxidase 1 n=2 Tax=Homo sapiens RepI... 51 3e-09
UniRef100_Q6GM76 MGC82107 protein n=1 Tax=Xenopus laevis RepID=Q... 54 3e-09
UniRef100_UPI0000E7F9C6 PREDICTED: hypothetical protein n=1 Tax=... 52 4e-09
UniRef100_UPI0000ECD379 Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAO... 52 4e-09
UniRef100_UPI00006D6D0A PREDICTED: similar to hydroxyacid oxidas... 50 4e-09
UniRef100_UPI000057F14F UPI000057F14F related cluster n=1 Tax=Bo... 50 4e-09
UniRef100_B0BNF9 Hydroxyacid oxidase 1 n=1 Tax=Rattus norvegicus... 50 4e-09
UniRef100_Q9WU19 Hydroxyacid oxidase 1 n=2 Tax=Mus musculus RepI... 50 4e-09
UniRef100_C1BKC4 Hydroxyacid oxidase 1 n=1 Tax=Osmerus mordax Re... 52 4e-09
UniRef100_A3SFF5 FMN-dependent alpha-hydroxy acid dehydrogenase ... 53 5e-09
UniRef100_A3SYV8 FMN-dependent alpha-hydroxy acid dehydrogenase ... 53 5e-09
UniRef100_UPI000185FCAF hypothetical protein BRAFLDRAFT_199392 n... 52 5e-09
UniRef100_Q5KIR0 Putative uncharacterized protein n=1 Tax=Filoba... 50 7e-09
UniRef100_B8NG63 FMN-dependent dehydrogenase family protein n=1 ... 52 1e-08
UniRef100_C5T9F4 FMN-dependent alpha-hydroxy acid dehydrogenase ... 50 1e-08
UniRef100_UPI000155E2F9 PREDICTED: similar to Hydroxyacid oxidas... 52 1e-08
UniRef100_Q2UE74 Glycolate oxidase n=1 Tax=Aspergillus oryzae Re... 52 1e-08
UniRef100_B5X381 Hydroxyacid oxidase 1 n=2 Tax=Salmo salar RepID... 50 1e-08
UniRef100_UPI000194B9FC PREDICTED: similar to MGC82107 protein i... 49 1e-08
UniRef100_UPI00017F06D4 PREDICTED: similar to Hydroxyacid oxidas... 51 1e-08
UniRef100_UPI000194B9FD PREDICTED: similar to MGC82107 protein i... 49 1e-08
UniRef100_UPI000151B05E hypothetical protein PGUG_04504 n=1 Tax=... 44 2e-08
UniRef100_A5DMK3 Putative uncharacterized protein n=1 Tax=Pichia... 44 2e-08
UniRef100_UPI000194BE7F PREDICTED: similar to hydroxyacid oxidas... 48 2e-08
UniRef100_UPI0000566FD8 PREDICTED: hypothetical protein n=1 Tax=... 50 2e-08
UniRef100_Q7SXX8 Hydroxyacid oxidase 2 (Long chain) n=1 Tax=Dani... 50 2e-08
UniRef100_B7FG12 Putative uncharacterized protein n=1 Tax=Medica... 62 2e-08
UniRef100_Q9NYQ3 Hydroxyacid oxidase 2 n=3 Tax=Homo sapiens RepI... 50 2e-08
UniRef100_C7ZDW2 Putative uncharacterized protein n=1 Tax=Nectri... 51 3e-08
UniRef100_UPI0000D99B2E PREDICTED: hydroxyacid oxidase 2 isoform... 50 3e-08
UniRef100_UPI0000D99B2D PREDICTED: hydroxyacid oxidase 2 isoform... 50 3e-08
UniRef100_C7YT35 Putative uncharacterized protein n=1 Tax=Nectri... 50 4e-08
UniRef100_Q82T12 Glycolate oxidase, (S)-2-hydroxy-acid oxidase, ... 57 5e-08
UniRef100_Q8JZR9 Hydroxyacid oxidase (Glycolate oxidase) 3 n=1 T... 50 5e-08
UniRef100_Q3UNU6 Putative uncharacterized protein n=1 Tax=Mus mu... 50 5e-08
UniRef100_Q9NYQ2 Hydroxyacid oxidase 2 n=2 Tax=Mus musculus RepI... 50 5e-08
UniRef100_Q587P0 Glycolate oxidase (Fragment) n=1 Tax=Fragaria x... 60 6e-08
UniRef100_A3IHB5 Glycolate oxidase n=1 Tax=Cyanothece sp. CCY011... 55 7e-08
UniRef100_A7HQ18 FMN-dependent alpha-hydroxy acid dehydrogenase ... 47 7e-08
UniRef100_B7RR92 Peroxisomal (S)-2-hydroxy-acid oxidase n=1 Tax=... 47 7e-08
UniRef100_Q3ZBW2 Hydroxyacid oxidase 2 n=1 Tax=Bos taurus RepID=... 49 7e-08
UniRef100_UPI0000EB296E Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAO... 50 9e-08
UniRef100_B7G7W1 Glycolate oxidase n=1 Tax=Phaeodactylum tricorn... 44 1e-07
UniRef100_C3Z3V2 Putative uncharacterized protein n=1 Tax=Branch... 50 1e-07
UniRef100_C3Z4C2 Putative uncharacterized protein n=1 Tax=Branch... 53 1e-07
UniRef100_B4VJJ4 FMN-dependent dehydrogenase superfamily n=1 Tax... 52 1e-07
UniRef100_B4J7J3 GH20058 n=1 Tax=Drosophila grimshawi RepID=B4J7... 54 1e-07
UniRef100_Q7MZC1 Similar to lactate oxidase n=1 Tax=Photorhabdus... 46 1e-07
UniRef100_A7REP0 Predicted protein n=1 Tax=Nematostella vectensi... 49 1e-07
UniRef100_Q9D2W7 Putative uncharacterized protein n=1 Tax=Mus mu... 50 1e-07
UniRef100_Q7QGT9 AGAP010885-PA (Fragment) n=1 Tax=Anopheles gamb... 59 2e-07
UniRef100_B1WYQ0 Probable FMN-dependent alpha-hydroxy acid dehyd... 52 2e-07
UniRef100_UPI00015B4BE8 PREDICTED: similar to (s)-2-hydroxy-acid... 54 2e-07
UniRef100_UPI00015B4BE0 PREDICTED: similar to (s)-2-hydroxy-acid... 50 2e-07
UniRef100_UPI000186A3E2 hypothetical protein BRAFLDRAFT_254568 n... 51 2e-07
UniRef100_B7QE37 (S)-2-hydroxy-acid oxidase, putative n=1 Tax=Ix... 44 2e-07
UniRef100_A7SBH2 Predicted protein n=1 Tax=Nematostella vectensi... 59 2e-07
UniRef100_A8IEL8 Glycolate oxidase n=1 Tax=Chlamydomonas reinhar... 52 2e-07
UniRef100_A1VQD5 FMN-dependent alpha-hydroxy acid dehydrogenase ... 48 3e-07
UniRef100_UPI000180B591 PREDICTED: similar to LOC100101335 prote... 48 3e-07
UniRef100_C3XVZ3 Putative uncharacterized protein n=1 Tax=Branch... 50 3e-07
UniRef100_B4LPJ5 GJ21929 n=1 Tax=Drosophila virilis RepID=B4LPJ5... 49 4e-07
UniRef100_Q17C65 (S)-2-hydroxy-acid oxidase n=1 Tax=Aedes aegypt... 52 5e-07
UniRef100_B4QAP7 GD10762 n=1 Tax=Drosophila simulans RepID=B4QAP... 50 5e-07
UniRef100_B4HN19 GM21244 n=1 Tax=Drosophila sechellia RepID=B4HN... 50 5e-07
UniRef100_B7Q493 Glycolate oxidase, putative (Fragment) n=1 Tax=... 42 5e-07
UniRef100_C7BNF5 Putative uncharacterized protein n=1 Tax=Photor... 44 7e-07
UniRef100_C7QGC6 FMN-dependent alpha-hydroxy acid dehydrogenase ... 45 7e-07
UniRef100_A6H8K0 LOC100101335 protein (Fragment) n=1 Tax=Xenopus... 45 7e-07
UniRef100_B4P7M9 GE12845 n=1 Tax=Drosophila yakuba RepID=B4P7M9_... 50 7e-07
UniRef100_B4LMS9 GJ21802 n=1 Tax=Drosophila virilis RepID=B4LMS9... 50 7e-07
UniRef100_C1DQ10 L-lactate dehydrogenase/FMN-dependent alpha-hyd... 45 1e-06
UniRef100_B9XKJ6 FMN-dependent alpha-hydroxy acid dehydrogenase ... 45 1e-06
UniRef100_C4GG06 Putative uncharacterized protein n=1 Tax=Kingel... 49 2e-06
UniRef100_Q28YL3 GA15579 n=1 Tax=Drosophila pseudoobscura pseudo... 51 2e-06
UniRef100_B4H8H1 GL20092 n=1 Tax=Drosophila persimilis RepID=B4H... 51 2e-06
UniRef100_B4MKB8 GK20637 n=1 Tax=Drosophila willistoni RepID=B4M... 51 2e-06
UniRef100_B4KNA0 GI18775 n=1 Tax=Drosophila mojavensis RepID=B4K... 50 2e-06
UniRef100_A1Z8D2 CG18003, isoform A n=1 Tax=Drosophila melanogas... 49 2e-06
UniRef100_B6IDX0 FI01464p (Fragment) n=1 Tax=Drosophila melanoga... 49 2e-06
UniRef100_A4SYM1 L-lactate dehydrogenase (Cytochrome) n=1 Tax=Po... 49 2e-06
UniRef100_B1GRK5 Putative uncharacterized protein n=1 Tax=Caenor... 43 2e-06
UniRef100_A1Z8D3 CG18003, isoform B n=1 Tax=Drosophila melanogas... 49 2e-06
UniRef100_C5SJU6 FMN-dependent alpha-hydroxy acid dehydrogenase ... 44 2e-06
UniRef100_Q86NM4 RH48327p n=1 Tax=Drosophila melanogaster RepID=... 49 2e-06
UniRef100_B0X407 Glycolate oxidase n=1 Tax=Culex quinquefasciatu... 46 2e-06
UniRef100_C0DX06 Putative uncharacterized protein n=1 Tax=Eikene... 49 3e-06
UniRef100_Q0RIC4 Putative FMN-dependent lactate dehydrogenase n=... 42 3e-06
UniRef100_UPI00005A4407 PREDICTED: similar to Hydroxyacid oxidas... 52 3e-06
UniRef100_B2J901 FMN-dependent alpha-hydroxy acid dehydrogenase ... 43 3e-06
UniRef100_B3NN34 GG20155 n=1 Tax=Drosophila erecta RepID=B3NN34_... 50 3e-06
UniRef100_UPI00015B4574 PREDICTED: similar to CG18003-PA n=1 Tax... 44 3e-06
UniRef100_UPI0001BB8E92 glycolate oxidase n=1 Tax=Acinetobacter ... 49 4e-06
UniRef100_UPI00015B4299 PREDICTED: similar to ENSANGP00000018221... 44 4e-06
UniRef100_Q07523 Hydroxyacid oxidase 2 n=1 Tax=Rattus norvegicus... 43 4e-06
UniRef100_Q6C9A7 YALI0D12661p n=1 Tax=Yarrowia lipolytica RepID=... 40 6e-06
UniRef100_A9E8E7 FMN-dependent alpha-hydroxy acid dehydrogenase ... 51 6e-06
UniRef100_B0X408 Peroxisomal n=1 Tax=Culex quinquefasciatus RepI... 54 7e-06
UniRef100_A9C3D8 FMN-dependent alpha-hydroxy acid dehydrogenase ... 50 7e-06
UniRef100_B1FTY1 L-lactate dehydrogenase (Cytochrome) n=1 Tax=Bu... 47 7e-06
UniRef100_A6CJ80 Isopentenyl-diphosphate delta-isomerase II 2 n=... 41 7e-06
UniRef100_Q1IWN3 (S)-2-hydroxy-acid oxidase n=1 Tax=Deinococcus ... 40 7e-06
UniRef100_UPI0000D56303 PREDICTED: similar to AGAP010885-PA n=1 ... 45 7e-06
UniRef100_UPI0000519D78 PREDICTED: similar to CG18003-PB, isofor... 41 7e-06
UniRef100_C1A4Y0 Glycolate oxidase n=1 Tax=Gemmatimonas aurantia... 41 7e-06
UniRef100_Q128S9 FMN-dependent alpha-hydroxy acid dehydrogenase ... 47 9e-06