[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB928350 RCE39545
(438 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
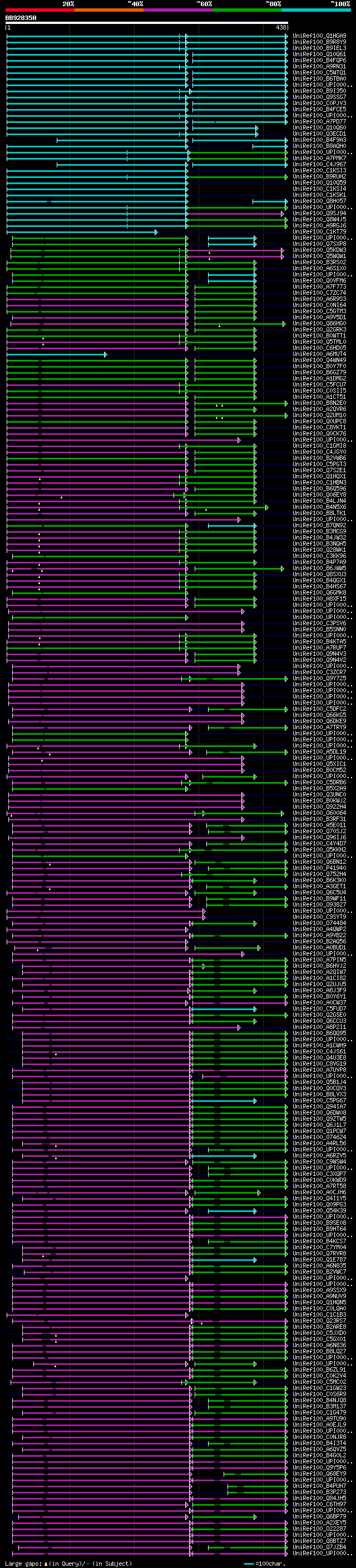
significant alignments:[graphical|details]
UniRef100_Q1HGA9 Putative GDP-mannose pyrophosphorylase n=1 Tax=... 166 3e-60
UniRef100_B9R8Y9 Mannose-1-phosphate guanyltransferase, putative... 167 1e-59
UniRef100_B9IEL3 Predicted protein n=1 Tax=Populus trichocarpa R... 160 4e-59
UniRef100_Q10Q61 Os03g0208900 protein n=1 Tax=Oryza sativa Japon... 163 4e-58
UniRef100_B4FQP6 Putative uncharacterized protein n=1 Tax=Zea ma... 160 1e-57
UniRef100_A9RN31 Predicted protein n=1 Tax=Physcomitrella patens... 151 4e-57
UniRef100_C5WTQ1 Putative uncharacterized protein Sb01g043370 n=... 159 4e-57
UniRef100_B6TBA0 Mannose-1-phosphate guanyltransferase n=1 Tax=Z... 158 6e-57
UniRef100_UPI0001985133 PREDICTED: hypothetical protein n=1 Tax=... 155 8e-57
UniRef100_B9I350 Predicted protein n=1 Tax=Populus trichocarpa R... 160 2e-56
UniRef100_Q9SSG7 F25A4.12 protein n=2 Tax=Arabidopsis thaliana R... 156 2e-56
UniRef100_C0PJV3 Putative uncharacterized protein n=1 Tax=Zea ma... 157 2e-55
UniRef100_B4FCE5 Putative uncharacterized protein n=1 Tax=Zea ma... 157 2e-55
UniRef100_UPI000162115C predicted protein n=1 Tax=Physcomitrella... 147 1e-54
UniRef100_A7PD77 Chromosome chr17 scaffold_12, whole genome shot... 155 1e-54
UniRef100_Q10Q60 ADP-glucose pyrophosphorylase family protein, p... 163 1e-50
UniRef100_Q3ECD1 Putative uncharacterized protein At1g74910.3 n=... 156 1e-49
UniRef100_B4F9A3 Putative uncharacterized protein n=1 Tax=Zea ma... 114 1e-43
UniRef100_B8AQH0 Putative uncharacterized protein n=1 Tax=Oryza ... 163 3e-42
UniRef100_UPI0001984B37 PREDICTED: hypothetical protein n=1 Tax=... 173 5e-42
UniRef100_A7PMK7 Chromosome chr14 scaffold_21, whole genome shot... 173 5e-42
UniRef100_C4J967 Putative uncharacterized protein n=1 Tax=Zea ma... 110 1e-41
UniRef100_C1KSI3 GMP1 (Fragment) n=1 Tax=Brachypodium distachyon... 164 2e-39
UniRef100_B9RUH2 Mannose-1-phosphate guanyltransferase, putative... 164 4e-39
UniRef100_Q10Q59 ADP-glucose pyrophosphorylase family protein, p... 163 5e-39
UniRef100_C1KSI4 GMP1 (Fragment) n=1 Tax=Brachypodium distachyon... 163 6e-39
UniRef100_C1KSK1 GMP1 (Fragment) n=1 Tax=Brachypodium distachyon... 162 1e-38
UniRef100_Q8H057 Putative GDP-mannose pyrophosphorylase n=1 Tax=... 147 2e-37
UniRef100_UPI00001622D9 ADP-glucose pyrophosphorylase family pro... 151 2e-35
UniRef100_Q9SJ94 Putative GDP-mannose pyrophosphorylase n=1 Tax=... 151 2e-35
UniRef100_Q8W4J5 Putative GDP-mannose pyrophosphorylase n=1 Tax=... 151 2e-35
UniRef100_A9RGJ6 Predicted protein n=1 Tax=Physcomitrella patens... 151 2e-35
UniRef100_C1KT79 GDP-mannose pyrophosphorylase (Fragment) n=1 Ta... 135 1e-30
UniRef100_UPI000188A0CD hypothetical protein LOC393469 n=1 Tax=D... 94 1e-22
UniRef100_Q7SXP8 Mannose-1-phosphate guanyltransferase alpha-B n... 94 1e-22
UniRef100_Q5KDW3 Mannose-1-phosphate guanylyltransferase, putati... 87 2e-20
UniRef100_Q5WQW1 Psa2p n=2 Tax=Filobasidiella neoformans RepID=Q... 87 2e-20
UniRef100_B3RS02 Putative uncharacterized protein n=1 Tax=Tricho... 78 2e-20
UniRef100_A6S1X0 Putative uncharacterized protein n=1 Tax=Botryo... 81 3e-20
UniRef100_UPI000069EA19 GDP-mannose pyrophosphorylase A n=1 Tax=... 85 4e-20
UniRef100_Q0VFM6 Mannose-1-phosphate guanyltransferase alpha n=1... 85 4e-20
UniRef100_A7F773 GDP-mannose pyrophosphorylase n=1 Tax=Sclerotin... 81 6e-20
UniRef100_C7ZC74 Predicted protein n=1 Tax=Nectria haematococca ... 82 1e-19
UniRef100_A6R9S3 Putative uncharacterized protein n=1 Tax=Ajello... 78 2e-19
UniRef100_C0NI64 GDP-mannose pyrophosphorylase A n=1 Tax=Ajellom... 78 2e-19
UniRef100_C5GTM3 GDP-mannose pyrophosphorylase A n=2 Tax=Ajellom... 77 3e-19
UniRef100_A9V5D1 Predicted protein n=1 Tax=Monosiga brevicollis ... 75 4e-19
UniRef100_Q86HG0 Mannose-1-phosphate guanyltransferase alpha n=1... 75 4e-19
UniRef100_Q2GRK3 Putative uncharacterized protein n=1 Tax=Chaeto... 79 5e-19
UniRef100_B0WTT1 Mannose-1-phosphate guanyltransferase n=1 Tax=C... 85 5e-19
UniRef100_Q5TML0 AGAP011723-PA n=1 Tax=Anopheles gambiae RepID=Q... 84 5e-19
UniRef100_C6HD05 GDP-mannose pyrophosphorylase A n=1 Tax=Ajellom... 78 6e-19
UniRef100_A6MUT4 GDP-mannose pyrophosphorylase (Fragment) n=1 Ta... 97 7e-19
UniRef100_Q4WN49 GDP-mannose pyrophosphorylase A n=1 Tax=Aspergi... 77 8e-19
UniRef100_B0Y7F0 GDP-mannose pyrophosphorylase A n=1 Tax=Aspergi... 77 8e-19
UniRef100_B6GZ79 Pc12g09190 protein n=1 Tax=Penicillium chrysoge... 78 1e-18
UniRef100_A1DMG2 GDP-mannose pyrophosphorylase A n=1 Tax=Neosart... 76 1e-18
UniRef100_C5FCU7 Mannose-1-phosphate guanyltransferase n=1 Tax=M... 76 1e-18
UniRef100_C0SII5 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 77 1e-18
UniRef100_A1CT51 GDP-mannose pyrophosphorylase A n=1 Tax=Aspergi... 77 1e-18
UniRef100_B8N2E0 GDP-mannose pyrophosphorylase A n=1 Tax=Aspergi... 74 2e-18
UniRef100_A2QVR6 Catalytic activity: GTP + alpha-D-Mannose 1-pho... 75 2e-18
UniRef100_Q2UM10 GDP-mannose pyrophosphorylase n=1 Tax=Aspergill... 74 2e-18
UniRef100_Q0UPC8 Putative uncharacterized protein n=1 Tax=Phaeos... 78 2e-18
UniRef100_C8VKT1 GDP-mannose pyrophosphorylase A (AFU_orthologue... 75 2e-18
UniRef100_Q0CK76 Putative uncharacterized protein n=1 Tax=Asperg... 75 3e-18
UniRef100_UPI0000E494C5 PREDICTED: hypothetical protein n=1 Tax=... 94 4e-18
UniRef100_C1GMI8 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 75 4e-18
UniRef100_C4JGY0 Putative uncharacterized protein n=1 Tax=Uncino... 75 5e-18
UniRef100_B2VWB6 Mannose-1-phosphate guanyltransferase 2 n=1 Tax... 77 5e-18
UniRef100_C5PGT3 Mannose-1-phosphate guanyltransferase, putative... 74 7e-18
UniRef100_Q7S2E1 Putative uncharacterized protein n=1 Tax=Neuros... 75 9e-18
UniRef100_Q1HQX1 GDP-mannose pyrophosphorylase A n=1 Tax=Aedes a... 80 9e-18
UniRef100_C1HBN3 Mannose-1-phosphate guanyltransferase subunit b... 75 1e-17
UniRef100_B6Q596 GDP-mannose pyrophosphorylase A n=1 Tax=Penicil... 72 1e-17
UniRef100_Q06EY8 GCD1 protein n=1 Tax=Terfezia boudieri RepID=Q0... 72 1e-17
UniRef100_B4LJN4 GJ21538 n=1 Tax=Drosophila virilis RepID=B4LJN4... 79 2e-17
UniRef100_B4N5X6 GK17960 n=1 Tax=Drosophila willistoni RepID=B4N... 78 2e-17
UniRef100_B8LTK1 GDP-mannose pyrophosphorylase A n=1 Tax=Talarom... 72 4e-17
UniRef100_UPI00006A452E PREDICTED: similar to MGC81801 protein n... 91 5e-17
UniRef100_B7QN92 GDP-mannose pyrophosphorylase, putative n=1 Tax... 82 6e-17
UniRef100_B3MCG9 GF13377 n=1 Tax=Drosophila ananassae RepID=B3MC... 79 7e-17
UniRef100_B4JW32 GH22966 n=1 Tax=Drosophila grimshawi RepID=B4JW... 77 7e-17
UniRef100_B3NQA5 GG22341 n=1 Tax=Drosophila erecta RepID=B3NQA5_... 79 9e-17
UniRef100_Q28WK1 GA20898 n=2 Tax=pseudoobscura subgroup RepID=Q2... 77 1e-16
UniRef100_C3KK96 Mannose-1-phosphate guanyltransferase alpha-A n... 89 2e-16
UniRef100_B4P7A9 GE14141 n=1 Tax=Drosophila yakuba RepID=B4P7A9_... 77 2e-16
UniRef100_B6JWW5 Mannose-1-phosphate guanyltransferase n=1 Tax=S... 71 2e-16
UniRef100_Q8SXU3 CG8207 n=1 Tax=Drosophila melanogaster RepID=Q8... 77 3e-16
UniRef100_B4QGX1 GD25606 n=1 Tax=Drosophila simulans RepID=B4QGX... 77 3e-16
UniRef100_B4HS67 GM20127 n=1 Tax=Drosophila sechellia RepID=B4HS... 77 3e-16
UniRef100_Q6GMK8 Mannose-1-phosphate guanyltransferase alpha-A n... 87 4e-16
UniRef100_A8XF15 Putative uncharacterized protein n=1 Tax=Caenor... 71 5e-16
UniRef100_UPI0000123FA7 Hypothetical protein CBG12186 n=1 Tax=Ca... 71 5e-16
UniRef100_UPI0001796223 PREDICTED: similar to GDP-mannose pyroph... 87 8e-16
UniRef100_UPI00017B3E9C UPI00017B3E9C related cluster n=1 Tax=Te... 87 8e-16
UniRef100_C3PSV6 GDP-mannose pyrophosphorylase A, isoform 2 (Pre... 87 8e-16
UniRef100_B5SNN0 GDP-mannose pyrophosphorylase A, isoform 1 (Pre... 86 1e-15
UniRef100_UPI0001926900 PREDICTED: similar to predicted protein ... 65 1e-15
UniRef100_B4KTA5 GI18453 n=1 Tax=Drosophila mojavensis RepID=B4K... 74 1e-15
UniRef100_A7RUP7 Predicted protein n=1 Tax=Nematostella vectensi... 70 1e-15
UniRef100_Q9N4V3 Putative uncharacterized protein n=1 Tax=Caenor... 69 1e-15
UniRef100_Q9N4V2 Putative uncharacterized protein n=1 Tax=Caenor... 69 1e-15
UniRef100_UPI00018651B3 hypothetical protein BRAFLDRAFT_88132 n=... 86 2e-15
UniRef100_C3ZCR7 Putative uncharacterized protein n=1 Tax=Branch... 86 2e-15
UniRef100_Q9Y725 Mannose-1-phosphate guanyltransferase 1 n=1 Tax... 66 2e-15
UniRef100_UPI00005BBCF6 PREDICTED: similar to GDP-mannose pyroph... 85 2e-15
UniRef100_UPI00005A5A4F PREDICTED: similar to GDP-mannose pyroph... 85 3e-15
UniRef100_UPI0000EB02DD GDP-mannose pyrophosphorylase A n=1 Tax=... 85 3e-15
UniRef100_UPI00004BFA83 PREDICTED: similar to GDP-mannose pyroph... 85 3e-15
UniRef100_C5DFC2 KLTH0D13948p n=1 Tax=Lachancea thermotolerans C... 66 4e-15
UniRef100_Q66KG5 Mannose-1-phosphate guanyltransferase alpha-B n... 84 4e-15
UniRef100_Q6DKE9 Mannose-1-phosphate guanyltransferase alpha-A n... 84 4e-15
UniRef100_A7TRY9 Putative uncharacterized protein n=1 Tax=Vander... 67 5e-15
UniRef100_UPI00016E5B7A UPI00016E5B7A related cluster n=1 Tax=Ta... 84 5e-15
UniRef100_UPI00016E5929 UPI00016E5929 related cluster n=1 Tax=Ta... 84 5e-15
UniRef100_UPI0000D57707 PREDICTED: similar to AGAP011723-PA n=1 ... 73 6e-15
UniRef100_A5DL19 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 67 6e-15
UniRef100_UPI00005A5A50 PREDICTED: similar to GDP-mannose pyroph... 83 8e-15
UniRef100_Q5XIC1 Mannose-1-phosphate guanyltransferase alpha n=1... 83 8e-15
UniRef100_B0CM52 Mannose-1-phosphate guanyltransferase alpha n=1... 83 8e-15
UniRef100_UPI0000DB779A PREDICTED: similar to CG8207-PA isoform ... 74 1e-14
UniRef100_C5DRB6 ZYRO0B07150p n=1 Tax=Zygosaccharomyces rouxii C... 69 1e-14
UniRef100_B5X2A9 Mannose-1-phosphate guanyltransferase alpha-A n... 83 1e-14
UniRef100_Q3UNC0 Putative uncharacterized protein (Fragment) n=1... 83 1e-14
UniRef100_B0KWJ2 GDP-mannose pyrophosphorylase A, isoform 2 (Pre... 83 1e-14
UniRef100_Q922H4 Mannose-1-phosphate guanyltransferase alpha n=1... 83 1e-14
UniRef100_O60064 Probable mannose-1-phosphate guanyltransferase ... 66 1e-14
UniRef100_B3RF31 Mannose-1-phosphate guanyltransferase alpha (Pr... 82 1e-14
UniRef100_A5E011 Mannose-1-phosphate guanyltransferase n=1 Tax=L... 66 2e-14
UniRef100_Q70SJ2 Mannose-1-phosphate guanyltransferase n=1 Tax=K... 64 2e-14
UniRef100_Q96IJ6 Mannose-1-phosphate guanyltransferase alpha n=1... 81 3e-14
UniRef100_C4Y4D7 Mannose-1-phosphate guanyltransferase n=1 Tax=C... 62 9e-14
UniRef100_Q5KKH2 Mannose-1-phosphate guanyltransferase n=2 Tax=F... 55 1e-13
UniRef100_UPI000194CBD1 PREDICTED: similar to GDP-mannose pyroph... 79 1e-13
UniRef100_Q6BN12 Mannose-1-phosphate guanyltransferase n=1 Tax=D... 61 1e-13
UniRef100_P41940 Mannose-1-phosphate guanyltransferase n=6 Tax=S... 64 1e-13
UniRef100_Q752H4 Mannose-1-phosphate guanyltransferase n=1 Tax=E... 65 2e-13
UniRef100_B6K3K0 Mannose-1-phosphate guanyltransferase Mpg1 n=1 ... 62 2e-13
UniRef100_A3GET1 Mannose-1-phosphate guanyltransferase (ATP-mann... 62 2e-13
UniRef100_Q6C5U4 YALI0E15125p n=1 Tax=Yarrowia lipolytica RepID=... 68 3e-13
UniRef100_B9WF11 Mannose-1-phosphate guanyltransferase, putative... 61 5e-13
UniRef100_O93827 Mannose-1-phosphate guanyltransferase n=1 Tax=C... 61 5e-13
UniRef100_UPI000186D94C glucose-1-phosphate adenylyltransferase,... 77 6e-13
UniRef100_C9SYT9 Mannose-1-phosphate guanyltransferase n=1 Tax=V... 77 8e-13
UniRef100_O74484 Mannose-1-phosphate guanyltransferase n=1 Tax=S... 61 9e-13
UniRef100_A4QWP2 Putative uncharacterized protein n=1 Tax=Magnap... 76 1e-12
UniRef100_A9VB22 Predicted protein n=1 Tax=Monosiga brevicollis ... 54 2e-12
UniRef100_B2AQ56 Predicted CDS Pa_4_5270 (Fragment) n=1 Tax=Podo... 75 2e-12
UniRef100_A0BUD1 Chromosome undetermined scaffold_129, whole gen... 60 4e-12
UniRef100_UPI0000F2E00D PREDICTED: similar to GDP-mannose pyroph... 74 5e-12
UniRef100_A7PIN5 Chromosome chr13 scaffold_17, whole genome shot... 54 6e-12
UniRef100_B6HVJ2 Pc22g06040 protein n=1 Tax=Penicillium chrysoge... 52 1e-11
UniRef100_A2QIW7 Catalytic activity: GTP + alpha-D-mannose 1-pho... 50 1e-11
UniRef100_A1CI82 Mannose-1-phosphate guanylyltransferase n=1 Tax... 53 2e-11
UniRef100_Q2UJU5 Mannose-1-phosphate guanyltransferase n=2 Tax=A... 50 2e-11
UniRef100_A8J3F9 GDP-D-mannose pyrophosphorylase n=1 Tax=Chlamyd... 57 2e-11
UniRef100_B0Y6Y1 Mannose-1-phosphate guanylyltransferase n=1 Tax... 52 2e-11
UniRef100_A0CW37 Chromosome undetermined scaffold_3, whole genom... 55 2e-11
UniRef100_C5FUD7 Mannose-1-phosphate guanyltransferase n=1 Tax=M... 52 3e-11
UniRef100_Q2GSE0 Putative uncharacterized protein n=1 Tax=Chaeto... 52 4e-11
UniRef100_Q6CCU3 Mannose-1-phosphate guanyltransferase n=1 Tax=Y... 54 4e-11
UniRef100_A8P2I1 Putative uncharacterized protein n=1 Tax=Brugia... 71 4e-11
UniRef100_B6QQ95 Mannose-1-phosphate guanylyltransferase n=1 Tax... 50 5e-11
UniRef100_UPI000051F513 mannose-1-phosphate guanylyltransferase ... 50 6e-11
UniRef100_A1CWH9 Mannose-1-phosphate guanylyltransferase n=1 Tax... 50 6e-11
UniRef100_C4JS61 Mannose-1-phosphate guanyltransferase n=1 Tax=U... 49 6e-11
UniRef100_Q4U3E8 Mannose-1-phosphate guanyltransferase n=1 Tax=A... 50 6e-11
UniRef100_C8VG19 Mannose-1-phosphate guanyltransferase (EC 2.7.7... 49 6e-11
UniRef100_A7UVP8 AGAP001299-PA n=1 Tax=Anopheles gambiae RepID=A... 57 6e-11
UniRef100_UPI000175844C PREDICTED: similar to mannose-1-phosphat... 54 6e-11
UniRef100_Q5B1J4 Mannose-1-phosphate guanyltransferase n=1 Tax=E... 49 6e-11
UniRef100_Q0CQV3 Mannose-1-phosphate guanyltransferase n=1 Tax=A... 50 6e-11
UniRef100_B8LVX3 Mannose-1-phosphate guanylyltransferase n=1 Tax... 49 8e-11
UniRef100_C5PG67 Mannose-1-phosphate guanyltransferase, putative... 49 1e-10
UniRef100_Q94IA7 GDP-D-mannose pyrophosphorylase n=1 Tax=Nicotia... 50 1e-10
UniRef100_Q6DW08 GMPase n=1 Tax=Medicago sativa RepID=Q6DW08_MEDSA 50 1e-10
UniRef100_Q9ZTW5 GDP-mannose pyrophosphorylase n=1 Tax=Solanum t... 50 2e-10
UniRef100_Q6J1L7 GDP-mannose pyrophosphorylase n=1 Tax=Solanum l... 49 2e-10
UniRef100_Q1PCW7 GDP-mannose pyrophosphorylase n=1 Tax=Solanum l... 49 2e-10
UniRef100_O74624 Mannose-1-phosphate guanyltransferase n=1 Tax=H... 49 3e-10
UniRef100_A4RL56 Putative uncharacterized protein n=1 Tax=Magnap... 47 3e-10
UniRef100_UPI00003C097A PREDICTED: similar to CG1129-PA, isoform... 52 3e-10
UniRef100_A6RZV5 Putative uncharacterized protein n=1 Tax=Botryo... 50 3e-10
UniRef100_C9WSW4 GDP-D-mannose pyrophosphorylase n=1 Tax=Glycine... 48 4e-10
UniRef100_UPI000186462D hypothetical protein BRAFLDRAFT_59509 n=... 56 4e-10
UniRef100_C3XQP7 Putative uncharacterized protein n=1 Tax=Branch... 56 4e-10
UniRef100_C0KWD9 GDP-D-mannose pyrophosphorylase n=1 Tax=Actinid... 48 5e-10
UniRef100_A7RT58 Predicted protein n=1 Tax=Nematostella vectensi... 49 5e-10
UniRef100_A0CJH6 Chromosome undetermined scaffold_2, whole genom... 57 6e-10
UniRef100_Q4I1Y5 Mannose-1-phosphate guanyltransferase n=1 Tax=G... 48 6e-10
UniRef100_Q09PG3 GDP-D-mannose pyrophosphorylase n=1 Tax=Viola b... 47 6e-10
UniRef100_Q54K39 Mannose-1-phosphate guanyltransferase beta n=1 ... 56 6e-10
UniRef100_UPI000194D311 PREDICTED: putative GDP-mannose pyrophos... 56 6e-10
UniRef100_B9SE08 Mannose-1-phosphate guanyltransferase, putative... 47 8e-10
UniRef100_B9HT64 Predicted protein n=1 Tax=Populus trichocarpa R... 47 8e-10
UniRef100_UPI0000E80E0B PREDICTED: similar to MGC84017 protein n... 56 1e-09
UniRef100_B4KCS7 GI10243 n=1 Tax=Drosophila mojavensis RepID=B4K... 52 1e-09
UniRef100_C7YM04 Predicted protein n=1 Tax=Nectria haematococca ... 48 1e-09
UniRef100_Q7RVR8 Mannose-1-phosphate guanyltransferase n=1 Tax=N... 47 1e-09
UniRef100_Q1E787 Putative uncharacterized protein n=1 Tax=Coccid... 46 1e-09
UniRef100_A6N835 GDP-mannose pyrophosphorylase n=1 Tax=Pinus tae... 46 1e-09
UniRef100_B2VWC7 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 49 1e-09
UniRef100_UPI000180C0FA PREDICTED: similar to GDP-mannose pyroph... 66 1e-09
UniRef100_UPI0000ECAC67 GDP-mannose pyrophosphorylase B isoform ... 56 1e-09
UniRef100_A9SSX9 Predicted protein n=1 Tax=Physcomitrella patens... 49 1e-09
UniRef100_A9NUV9 Putative uncharacterized protein n=1 Tax=Picea ... 47 1e-09
UniRef100_Q1HQN5 GDP-mannose pyrophosphorylase B n=1 Tax=Aedes a... 55 1e-09
UniRef100_C0LQA0 GDP-D-mannose pyrophosphorylase (Fragment) n=1 ... 47 1e-09
UniRef100_C1C1B3 Mannose-1-phosphate guanyltransferase alpha-A n... 66 1e-09
UniRef100_Q23RS7 Nucleotidyl transferase family protein n=1 Tax=... 51 2e-09
UniRef100_B2ARE8 Predicted CDS Pa_4_7730 n=1 Tax=Podospora anser... 47 2e-09
UniRef100_C5JXD0 Mannose-1-phosphate guanylyltransferase n=1 Tax... 45 2e-09
UniRef100_C5GX01 Mannose-1-phosphate guanylyltransferase n=1 Tax... 45 2e-09
UniRef100_A6N836 GDP-mannose pyrophosphorylase n=1 Tax=Pinus tae... 48 2e-09
UniRef100_B8LQ27 Putative uncharacterized protein n=1 Tax=Picea ... 45 2e-09
UniRef100_UPI000192500E PREDICTED: similar to predicted protein,... 45 2e-09
UniRef100_UPI0001791D0A PREDICTED: similar to GA20898-PA n=1 Tax... 57 2e-09
UniRef100_B6ZL91 GDP-D-mannose pyrophosphorylase n=1 Tax=Prunus ... 46 2e-09
UniRef100_C0K2V4 GDP-D-mannose pyrophosphorylase (Fragment) n=1 ... 47 2e-09
UniRef100_C5MC02 Putative uncharacterized protein n=1 Tax=Candid... 53 3e-09
UniRef100_C1GW23 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 47 3e-09
UniRef100_C0S6R9 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 47 3e-09
UniRef100_B4NJQ8 GK12822 n=1 Tax=Drosophila willistoni RepID=B4N... 50 3e-09
UniRef100_B3M137 GF18913 n=1 Tax=Drosophila ananassae RepID=B3M1... 50 3e-09
UniRef100_C1G479 Mannose-1-phosphate guanyltransferase n=1 Tax=P... 47 3e-09
UniRef100_A9TU90 Predicted protein n=1 Tax=Physcomitrella patens... 48 3e-09
UniRef100_A0EJL9 GDP-mannose pyrophosphorylase n=1 Tax=Malpighia... 45 3e-09
UniRef100_UPI00005A3AF7 PREDICTED: similar to GDP-mannose pyroph... 54 4e-09
UniRef100_C0NJR8 Mannose-1-phosphate guanylyltransferase n=1 Tax... 44 4e-09
UniRef100_B4I3T4 GM10777 n=1 Tax=Drosophila sechellia RepID=B4I3... 50 4e-09
UniRef100_A6QVZ5 Mannose-1-phosphate guanyltransferase n=1 Tax=A... 44 4e-09
UniRef100_B4G0L2 Putative uncharacterized protein n=1 Tax=Zea ma... 46 4e-09
UniRef100_UPI00005A3AF6 PREDICTED: similar to GDP-mannose pyroph... 54 4e-09
UniRef100_Q9Y5P6 Mannose-1-phosphate guanyltransferase beta n=1 ... 54 4e-09
UniRef100_Q68EY9 Mannose-1-phosphate guanyltransferase beta-A n=... 53 4e-09
UniRef100_UPI000179D375 hypothetical protein LOC514161 n=1 Tax=B... 53 5e-09
UniRef100_B4PUH7 GE25436 n=1 Tax=Drosophila yakuba RepID=B4PUH7_... 50 6e-09
UniRef100_B3P273 GG12578 n=1 Tax=Drosophila erecta RepID=B3P273_... 50 6e-09
UniRef100_Q84JH5 Os03g0268400 protein n=1 Tax=Oryza sativa Japon... 47 6e-09
UniRef100_C6TH97 Putative uncharacterized protein n=1 Tax=Glycin... 44 6e-09
UniRef100_UPI00017972C7 PREDICTED: similar to GDP-mannose pyroph... 53 7e-09
UniRef100_Q6BP79 DEHA2E15862p n=2 Tax=Debaryomyces hansenii RepI... 48 8e-09
UniRef100_A2XEY5 Putative uncharacterized protein n=1 Tax=Oryza ... 46 8e-09
UniRef100_O22287 GDP-mannose pyrophosphorylase n=1 Tax=Arabidops... 44 8e-09
UniRef100_UPI00006D4FDE PREDICTED: similar to GDP-mannose pyroph... 52 8e-09
UniRef100_Q8BTZ7 Mannose-1-phosphate guanyltransferase beta n=1 ... 52 8e-09
UniRef100_Q7JZB4 Mannose-1-phosphate guanyltransferase beta n=1 ... 48 1e-08
UniRef100_UPI0000250FB9 GDP-mannose pyrophosphorylase B n=1 Tax=... 52 1e-08
UniRef100_C9ST00 Mannose-1-phosphate guanyltransferase n=1 Tax=V... 45 1e-08
UniRef100_Q295Y7 Mannose-1-phosphate guanyltransferase beta n=2 ... 47 2e-08
UniRef100_Q2YDJ9 Mannose-1-phosphate guanyltransferase beta n=1 ... 51 2e-08
UniRef100_Q68EQ1 Mannose-1-phosphate guanyltransferase beta n=1 ... 50 2e-08
UniRef100_UPI0001A2BF05 hypothetical protein LOC445097 n=1 Tax=D... 50 2e-08
UniRef100_Q6DBU5 Mannose-1-phosphate guanyltransferase beta n=1 ... 50 2e-08
UniRef100_B9HK47 Predicted protein n=1 Tax=Populus trichocarpa R... 50 2e-08
UniRef100_B4JSV2 GH23007 n=1 Tax=Drosophila grimshawi RepID=B4JS... 47 2e-08
UniRef100_Q9M2S0 Mannose-1-phosphate guanylyltransferase-like pr... 46 2e-08
UniRef100_A2VD83 Mannose-1-phosphate guanyltransferase beta-B n=... 50 2e-08
UniRef100_Q6FRY2 Mannose-1-phosphate guanyltransferase 2 n=1 Tax... 62 3e-08
UniRef100_B9W9U1 Mannose-1-phosphate guanyltransferase, putative... 46 3e-08
UniRef100_C4QZM0 GDP-mannose pyrophosphorylase (Mannose-1-phosph... 53 3e-08
UniRef100_C4QA95 Mannose-1-phosphate guanyltransferase, putative... 61 3e-08
UniRef100_P0C5I2 Mannose-1-phosphate guanyltransferase beta n=1 ... 54 4e-08
UniRef100_C6TGC7 Putative uncharacterized protein (Fragment) n=1... 49 4e-08
UniRef100_Q5AL34 Putative uncharacterized protein n=1 Tax=Candid... 45 5e-08
UniRef100_Q9C5B8 GDP-mannose pyrophosphorylase n=1 Tax=Arabidops... 41 5e-08
UniRef100_UPI000036B54D PREDICTED: GDP-mannose pyrophosphorylase... 54 9e-08
UniRef100_Q9Y5P6-2 Isoform 2 of Mannose-1-phosphate guanyltransf... 54 9e-08
UniRef100_UPI000179320E PREDICTED: similar to Mannose-1-phosphat... 53 1e-07
UniRef100_C5KTB9 Mannose-1-phosphate guanyltransferase, putative... 40 1e-07
UniRef100_A9NVX8 Putative uncharacterized protein n=1 Tax=Picea ... 42 1e-07
UniRef100_C4PX01 Gdp-mannose pyrophosphorylase b, isoform 2 n=1 ... 46 2e-07
UniRef100_UPI00006D4FDD PREDICTED: similar to GDP-mannose pyroph... 52 2e-07
UniRef100_A3GHU9 Probable mannose-1-phosphate guanyltransferase ... 50 3e-07
UniRef100_C5LME3 Mannose-1-phosphate guanyltransferase, putative... 42 4e-07
UniRef100_C4Q516 Glucosamine-1-phosphate N-acetyltransferase n=1... 47 5e-07
UniRef100_UPI00006609C9 UPI00006609C9 related cluster n=1 Tax=Ta... 57 8e-07
UniRef100_A4HCM4 Mannose-1-phosphate guanyltransferase n=1 Tax=L... 41 9e-07
UniRef100_C5M0M8 Mannose-1-phosphate guanyltransferase, putative... 42 9e-07
UniRef100_A0DQV4 Chromosome undetermined scaffold_6, whole genom... 56 1e-06
UniRef100_UPI00015B44C6 PREDICTED: similar to GA10892-PA n=1 Tax... 42 2e-06
UniRef100_B9PL80 Mannose-1-phosphate guanylyltransferase, putati... 43 3e-06
UniRef100_B6KB36 Mannose-1-phosphate guanylyltransferase, putati... 43 3e-06
UniRef100_A8Q0Z0 GDP-mannose pyrophosphorylase B, isoform 2, put... 46 3e-06
UniRef100_B4M536 GJ11048 n=1 Tax=Drosophila virilis RepID=B4M536... 54 5e-06
UniRef100_B7PWY1 GDP-mannose pyrophosphorylase/mannose-1-phospha... 45 7e-06
UniRef100_B4QVM8 GD19750 n=1 Tax=Drosophila simulans RepID=B4QVM... 39 8e-06
UniRef100_Q5CHS1 GDP-mannose pyrophosphorylase (4N40) n=1 Tax=Cr... 41 9e-06