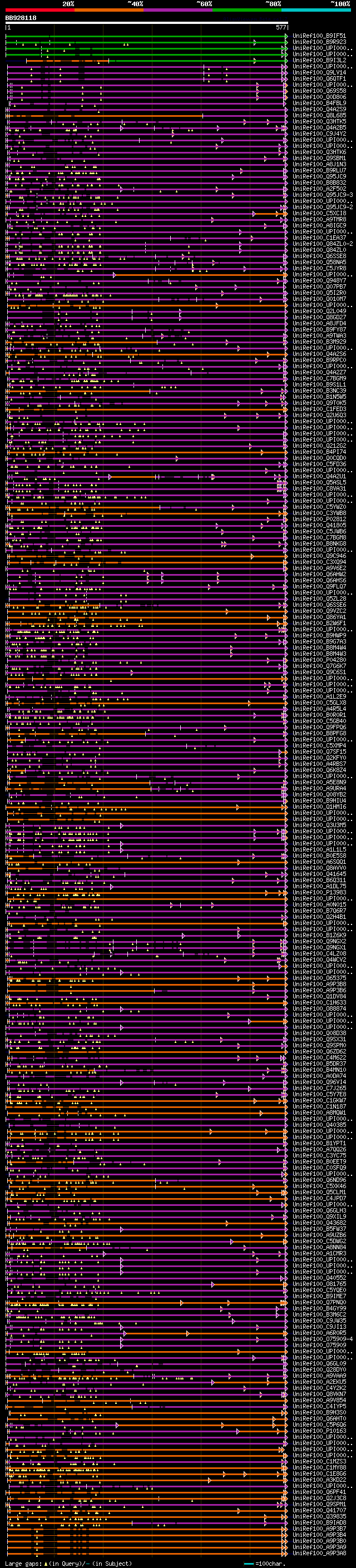
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB928118 RCE39276
(577 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9IF51 Predicted protein n=1 Tax=Populus trichocarpa R... 259 1e-67
UniRef100_B9R923 Epstein-Barr nuclear antigen, putative n=1 Tax=... 254 3e-66
UniRef100_UPI0001985297 PREDICTED: hypothetical protein isoform ... 226 1e-57
UniRef100_UPI0001985296 PREDICTED: hypothetical protein isoform ... 226 1e-57
UniRef100_B9I3L2 Predicted protein n=1 Tax=Populus trichocarpa R... 182 2e-44
UniRef100_UPI0001A7B301 proline-rich family protein n=1 Tax=Arab... 168 3e-40
UniRef100_Q9LV14 Genomic DNA, chromosome 5, P1 clone:MRG21 n=1 T... 168 3e-40
UniRef100_Q6QTF1 EARLY FLOWERING 5 n=1 Tax=Arabidopsis thaliana ... 168 3e-40
UniRef100_UPI0000E12A1F Os07g0192900 n=1 Tax=Oryza sativa Japoni... 157 4e-37
UniRef100_Q69S58 Putative hydroxyproline-rich glycoprotein DZ-HR... 157 4e-37
UniRef100_Q0D806 Os07g0192900 protein n=1 Tax=Oryza sativa Japon... 157 4e-37
UniRef100_B4FBL9 Putative uncharacterized protein n=1 Tax=Zea ma... 154 6e-36
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 128 4e-28
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 127 6e-28
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 127 8e-28
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 125 2e-27
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 125 3e-27
UniRef100_UPI0001982977 PREDICTED: hypothetical protein n=1 Tax=... 124 4e-27
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 122 2e-26
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 122 2e-26
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 122 3e-26
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 121 3e-26
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 121 4e-26
UniRef100_Q95JC9 Parotid hormone n=1 Tax=Sus scrofa RepID=PRP_PIG 120 6e-26
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 120 1e-25
UniRef100_A2F502 Formin Homology 2 Domain containing protein n=1... 119 2e-25
UniRef100_Q95JC9-3 Isoform 3 of Basic proline-rich protein n=1 T... 118 3e-25
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 118 4e-25
UniRef100_Q95JC9-2 Isoform 2 of Basic proline-rich protein n=1 T... 118 4e-25
UniRef100_C5XCI8 Putative uncharacterized protein Sb02g005690 n=... 117 6e-25
UniRef100_A9TMR8 Predicted protein n=1 Tax=Physcomitrella patens... 116 1e-24
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 115 2e-24
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 115 2e-24
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 115 2e-24
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 115 2e-24
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 115 2e-24
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 115 3e-24
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 114 4e-24
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 114 4e-24
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 114 7e-24
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 114 7e-24
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 113 9e-24
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 113 1e-23
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 113 1e-23
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 112 2e-23
UniRef100_Q2L049 Filamentous hemagglutinin/adhesin n=1 Tax=Borde... 112 2e-23
UniRef100_Q8GD27 Adhesin FhaB n=1 Tax=Bordetella avium RepID=Q8G... 112 2e-23
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 112 3e-23
UniRef100_B9FY87 Putative uncharacterized protein n=1 Tax=Oryza ... 111 4e-23
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 111 4e-23
UniRef100_B3M929 GF25073 n=1 Tax=Drosophila ananassae RepID=B3M9... 111 5e-23
UniRef100_UPI0000E800CF PREDICTED: similar to formin 2 n=1 Tax=G... 110 6e-23
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 110 6e-23
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 110 6e-23
UniRef100_UPI0000DB7674 PREDICTED: hypothetical protein n=1 Tax=... 110 8e-23
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 110 1e-22
UniRef100_C7BGM9 Formin 2B n=1 Tax=Physcomitrella patens RepID=C... 110 1e-22
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 110 1e-22
UniRef100_B3NC39 GG15230 n=1 Tax=Drosophila erecta RepID=B3NC39_... 110 1e-22
UniRef100_B1N5W5 Diaphanous protein, putative (Fragment) n=1 Tax... 109 2e-22
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 108 2e-22
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 108 2e-22
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 108 4e-22
UniRef100_UPI0001983015 PREDICTED: hypothetical protein n=1 Tax=... 107 5e-22
UniRef100_UPI0000ECC8C2 Formin-2. n=1 Tax=Gallus gallus RepID=UP... 107 5e-22
UniRef100_UPI0000E23041 PREDICTED: hypothetical protein isoform ... 107 7e-22
UniRef100_UPI0000E23040 PREDICTED: hypothetical protein isoform ... 107 7e-22
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 107 7e-22
UniRef100_B4PI74 GE21449 n=1 Tax=Drosophila yakuba RepID=B4PI74_... 107 7e-22
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 107 7e-22
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 107 7e-22
UniRef100_UPI000023F096 hypothetical protein FG08656.1 n=1 Tax=G... 106 1e-21
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 106 1e-21
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 106 1e-21
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 106 1e-21
UniRef100_UPI0000E48E71 PREDICTED: similar to Cpsf6 protein, par... 106 1e-21
UniRef100_UPI0000EE5993 proline-rich protein BstNI subfamily 2 n... 106 1e-21
UniRef100_C5YWZ0 Putative uncharacterized protein Sb09g030460 n=... 106 1e-21
UniRef100_C3YWB8 Putative uncharacterized protein n=1 Tax=Branch... 106 1e-21
UniRef100_P02812 Basic proline-rich peptide IB-4 n=1 Tax=Homo sa... 106 1e-21
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 105 2e-21
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 105 2e-21
UniRef100_C7BGM8 Formin 2A n=1 Tax=Physcomitrella patens RepID=C... 105 3e-21
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 105 3e-21
UniRef100_UPI00017600C0 PREDICTED: similar to abl-interactor 1 n... 105 3e-21
UniRef100_Q9C946 Putative uncharacterized protein T7P1.21 n=1 Ta... 105 3e-21
UniRef100_C3XQ94 Putative uncharacterized protein n=1 Tax=Branch... 105 3e-21
UniRef100_A9V6E2 Predicted protein n=1 Tax=Monosiga brevicollis ... 105 3e-21
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 105 3e-21
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 105 3e-21
UniRef100_Q9FLQ7 Formin-like protein 20 n=1 Tax=Arabidopsis thal... 105 3e-21
UniRef100_UPI0000611322 cyclin K n=1 Tax=Gallus gallus RepID=UPI... 103 7e-21
UniRef100_Q5ZL28 Putative uncharacterized protein n=1 Tax=Gallus... 103 7e-21
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 103 7e-21
UniRef100_Q9VZC2 CG15021 n=1 Tax=Drosophila melanogaster RepID=Q... 103 7e-21
UniRef100_Q86YA1 PRB1 protein (Fragment) n=1 Tax=Homo sapiens Re... 103 7e-21
UniRef100_B2W6F1 Putative uncharacterized protein n=1 Tax=Pyreno... 103 7e-21
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 103 1e-20
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 103 1e-20
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 103 1e-20
UniRef100_B8M4W4 Actin associated protein Wsp1, putative n=1 Tax... 103 1e-20
UniRef100_B8M4W3 Actin associated protein Wsp1, putative n=1 Tax... 103 1e-20
UniRef100_P04280 Peptide P-H n=1 Tax=Homo sapiens RepID=PRP1_HUMAN 103 1e-20
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 103 1e-20
UniRef100_Q9C6S1 Formin-like protein 14 n=1 Tax=Arabidopsis thal... 103 1e-20
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 103 1e-20
UniRef100_UPI0000F2BCA5 PREDICTED: similar to formin 2 n=1 Tax=M... 103 1e-20
UniRef100_UPI00002371B7 proline-rich protein BstNI subfamily 1 i... 103 1e-20
UniRef100_A1L2E9 LOC566059 protein (Fragment) n=1 Tax=Danio reri... 103 1e-20
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 103 1e-20
UniRef100_A4R5L4 Putative uncharacterized protein n=1 Tax=Magnap... 103 1e-20
UniRef100_B0R0R1 Novel protein similar to human Ras association ... 102 2e-20
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 102 2e-20
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 102 2e-20
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 102 2e-20
UniRef100_UPI0000DF08BC Os02g0794900 n=1 Tax=Oryza sativa Japoni... 102 3e-20
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 102 3e-20
UniRef100_Q7SF15 WASP-like pretein las17p n=1 Tax=Neurospora cra... 102 3e-20
UniRef100_Q2KFY0 Putative uncharacterized protein n=1 Tax=Magnap... 102 3e-20
UniRef100_A4RBS7 Putative uncharacterized protein n=1 Tax=Magnap... 102 3e-20
UniRef100_Q6K8Z4 Formin-like protein 7 n=1 Tax=Oryza sativa Japo... 102 3e-20
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 101 4e-20
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 101 4e-20
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 101 4e-20
UniRef100_Q08YB2 Response regulator n=1 Tax=Stigmatella aurantia... 101 5e-20
UniRef100_B9HIU4 Predicted protein (Fragment) n=1 Tax=Populus tr... 101 5e-20
UniRef100_Q1HMI6 Formin A n=2 Tax=Trypanosoma cruzi RepID=Q1HMI6... 101 5e-20
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 100 6e-20
UniRef100_UPI00006A03ED MGC69562 protein. n=1 Tax=Xenopus (Silur... 100 6e-20
UniRef100_Q3U3M5 Putative uncharacterized protein n=1 Tax=Mus mu... 100 6e-20
UniRef100_UPI000175F21B PREDICTED: similar to Ras-associated and... 100 8e-20
UniRef100_UPI00015A6C38 UPI00015A6C38 related cluster n=1 Tax=Da... 100 8e-20
UniRef100_UPI0001B7AFE7 UPI0001B7AFE7 related cluster n=1 Tax=Ra... 100 8e-20
UniRef100_A1L1L5 Ccnk protein n=1 Tax=Rattus norvegicus RepID=A1... 100 8e-20
UniRef100_B0E5S8 Formin 2,3 and collagen domain-containing prote... 100 8e-20
UniRef100_A6SQQ1 Putative uncharacterized protein n=1 Tax=Botryo... 100 8e-20
UniRef100_Q8AVV0 Sf3a2-prov protein n=1 Tax=Xenopus laevis RepID... 100 1e-19
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 100 1e-19
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 100 1e-19
UniRef100_A1DL75 Actin associated protein Wsp1, putative n=1 Tax... 100 1e-19
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 100 1e-19
UniRef100_UPI000155BA60 PREDICTED: similar to hCG2029577, partia... 99 2e-19
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 99 2e-19
UniRef100_B7Q6R7 Spliceosome associated protein, putative n=1 Ta... 99 2e-19
UniRef100_Q2H4B1 Putative uncharacterized protein n=1 Tax=Chaeto... 99 2e-19
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 99 2e-19
UniRef100_UPI00006A03EC MGC69562 protein. n=1 Tax=Xenopus (Silur... 99 2e-19
UniRef100_B1Z6K9 Type VI secretion system Vgr family protein n=1... 99 2e-19
UniRef100_Q9NGX2 Diaphanous protein n=1 Tax=Entamoeba histolytic... 99 2e-19
UniRef100_Q9NGX1 Diaphanous protein (Fragment) n=1 Tax=Entamoeba... 99 2e-19
UniRef100_C4LZ08 Diaphanous protein, homolog 1, putative n=1 Tax... 99 2e-19
UniRef100_Q4WCV2 Actin associated protein Wsp1, putative n=1 Tax... 99 2e-19
UniRef100_UPI000184A471 Formin-2. n=1 Tax=Xenopus (Silurana) tro... 99 3e-19
UniRef100_UPI00001C4FB6 cyclin K n=1 Tax=Mus musculus RepID=UPI0... 99 3e-19
UniRef100_O65375 F12F1.9 protein n=1 Tax=Arabidopsis thaliana Re... 99 3e-19
UniRef100_A9P3B8 Anther-specific proline rich protein n=1 Tax=Br... 99 3e-19
UniRef100_A9P3B6 Anther-specific proline rich protein n=1 Tax=Br... 99 3e-19
UniRef100_Q1DV84 Putative uncharacterized protein n=1 Tax=Coccid... 99 3e-19
UniRef100_C1H633 Predicted protein n=1 Tax=Paracoccidioides bras... 99 3e-19
UniRef100_O88874 Cyclin-K n=1 Tax=Mus musculus RepID=CCNK_MOUSE 99 3e-19
UniRef100_UPI000194C8D1 PREDICTED: cyclin K n=1 Tax=Taeniopygia ... 98 4e-19
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 98 4e-19
UniRef100_UPI0001B7AFE8 similar to cyclin K (LOC500715), mRNA n=... 98 4e-19
UniRef100_Q08D38 LOC779576 protein (Fragment) n=1 Tax=Xenopus (S... 98 4e-19
UniRef100_Q9SX31 F24J5.8 protein n=1 Tax=Arabidopsis thaliana Re... 98 4e-19
UniRef100_Q9SPM0 Extensin-like protein n=1 Tax=Zea mays RepID=Q9... 98 4e-19
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 98 4e-19
UniRef100_C4M622 Diaphanous protein, putative n=1 Tax=Entamoeba ... 98 4e-19
UniRef100_B5DR37 GA28529 n=1 Tax=Drosophila pseudoobscura pseudo... 98 4e-19
UniRef100_B4MN10 GK16584 n=1 Tax=Drosophila willistoni RepID=B4M... 98 4e-19
UniRef100_A0DA74 Chromosome undetermined scaffold_43, whole geno... 98 4e-19
UniRef100_Q96VI4 Protease 1 n=1 Tax=Pneumocystis carinii RepID=Q... 98 4e-19
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 98 5e-19
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 98 5e-19
UniRef100_C1GKW7 Predicted protein n=1 Tax=Paracoccidioides bras... 98 5e-19
UniRef100_C1N187 Putative uncharacterized protein n=1 Tax=Microm... 97 7e-19
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 97 7e-19
UniRef100_UPI0000DC195F UPI0000DC195F related cluster n=1 Tax=Ra... 97 9e-19
UniRef100_Q40385 Pistil extensin-like protein n=1 Tax=Nicotiana ... 97 9e-19
UniRef100_UPI0000E4A721 PREDICTED: similar to LOC397922 protein ... 97 1e-18
UniRef100_UPI0000E48217 PREDICTED: similar to LOC397922 protein,... 97 1e-18
UniRef100_B1YPT1 Type VI secretion system Vgr family protein n=1... 97 1e-18
UniRef100_A7QQ26 Chromosome chr2 scaffold_140, whole genome shot... 97 1e-18
UniRef100_C3YC75 Putative uncharacterized protein n=1 Tax=Branch... 97 1e-18
UniRef100_B0EET9 Putative uncharacterized protein n=1 Tax=Entamo... 97 1e-18
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 97 1e-18
UniRef100_UPI0001A2C970 WAS/WASL interacting protein family, mem... 96 2e-18
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 96 2e-18
UniRef100_C5XK46 Putative uncharacterized protein Sb03g034710 n=... 96 2e-18
UniRef100_Q5CLM1 Putative uncharacterized protein n=1 Tax=Crypto... 96 2e-18
UniRef100_C4JPD7 Putative uncharacterized protein n=1 Tax=Uncino... 96 2e-18
UniRef100_UPI00016E324C UPI00016E324C related cluster n=1 Tax=Ta... 96 2e-18
UniRef100_Q6GLH3 Splicing factor 3a, subunit 2, 66kDa n=1 Tax=Xe... 96 2e-18
UniRef100_Q9XIL9 Putative uncharacterized protein At2g15880 n=1 ... 96 2e-18
UniRef100_Q43682 Extensin-like protein (Fragment) n=1 Tax=Vigna ... 96 2e-18
UniRef100_B5FW37 Cyclin K isoform 1 (Predicted) n=1 Tax=Otolemur... 96 2e-18
UniRef100_A9UZB6 Predicted protein n=1 Tax=Monosiga brevicollis ... 96 2e-18
UniRef100_C5DWG2 ZYRO0D14586p n=1 Tax=Zygosaccharomyces rouxii C... 96 2e-18
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 96 2e-18
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 96 2e-18
UniRef100_UPI00017C39ED PREDICTED: similar to cyclin K n=1 Tax=B... 96 3e-18
UniRef100_UPI0000D9BD9F PREDICTED: similar to cyclin K n=1 Tax=M... 96 3e-18
UniRef100_UPI000179D978 UPI000179D978 related cluster n=1 Tax=Bo... 96 3e-18
UniRef100_Q40552 Pistil extensin like protein (Fragment) n=1 Tax... 96 3e-18
UniRef100_O81765 Extensin-like protein n=1 Tax=Arabidopsis thali... 96 3e-18
UniRef100_C5YQE0 Putative uncharacterized protein Sb08g019190 n=... 96 3e-18
UniRef100_B9IME7 Predicted protein (Fragment) n=1 Tax=Populus tr... 96 3e-18
UniRef100_Q7PNQ0 AGAP005952-PA n=1 Tax=Anopheles gambiae RepID=Q... 96 3e-18
UniRef100_B4GY99 GL19880 n=1 Tax=Drosophila persimilis RepID=B4G... 96 3e-18
UniRef100_B3M6C2 GF24314 n=1 Tax=Drosophila ananassae RepID=B3M6... 96 3e-18
UniRef100_C9JW35 Putative uncharacterized protein PRB3 n=1 Tax=H... 96 3e-18
UniRef100_C9JI13 Putative uncharacterized protein CCNK n=1 Tax=H... 96 3e-18
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 96 3e-18
UniRef100_O75909-4 Isoform 4 of Cyclin-K n=1 Tax=Homo sapiens Re... 96 3e-18
UniRef100_O75909 Cyclin-K n=1 Tax=Homo sapiens RepID=CCNK_HUMAN 96 3e-18
UniRef100_UPI000175FBE6 PREDICTED: similar to Wiskott-Aldrich sy... 95 3e-18
UniRef100_UPI0000223553 Hypothetical protein CBG15933 n=1 Tax=Ca... 95 3e-18
UniRef100_Q6GL09 WW domain binding protein 11 n=1 Tax=Xenopus (S... 95 3e-18
UniRef100_Q28DY0 WW domain binding protein 11 n=1 Tax=Xenopus (S... 95 3e-18
UniRef100_A9VAA9 Predicted protein n=1 Tax=Monosiga brevicollis ... 95 3e-18
UniRef100_A2EKU5 Putative uncharacterized protein n=1 Tax=Tricho... 95 3e-18
UniRef100_C4Y2K2 Putative uncharacterized protein n=1 Tax=Clavis... 95 3e-18
UniRef100_Q8VKN7 Putative uncharacterized protein n=1 Tax=Mycoba... 95 4e-18
UniRef100_A9V854 Predicted protein n=1 Tax=Monosiga brevicollis ... 95 4e-18
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 94 6e-18
UniRef100_B9H3S0 Predicted protein (Fragment) n=1 Tax=Populus tr... 94 6e-18
UniRef100_Q6AHT0 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 94 6e-18
UniRef100_C5P6Q6 WH1 domain containing protein n=1 Tax=Coccidioi... 94 6e-18
UniRef100_P10163 Peptide P-D n=1 Tax=Homo sapiens RepID=PRB4_HUMAN 94 6e-18
UniRef100_UPI000175FEF9 PREDICTED: hypothetical protein n=1 Tax=... 94 8e-18
UniRef100_UPI0000F20703 PREDICTED: similar to SH3 domain binding... 94 8e-18
UniRef100_UPI0000E80902 PREDICTED: similar to RAPH1 protein n=1 ... 94 8e-18
UniRef100_UPI0000546264 splicing factor 1 n=1 Tax=Danio rerio Re... 94 8e-18
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 94 8e-18
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 94 8e-18
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 94 8e-18
UniRef100_A3KD22 Leucine-rich repeat/extensin 1 n=1 Tax=Nicotian... 94 8e-18
UniRef100_UPI0000ECBAD6 inverted formin 2 isoform 1 n=1 Tax=Gall... 94 1e-17
UniRef100_Q6PF41 MGC68961 protein n=1 Tax=Xenopus laevis RepID=Q... 94 1e-17
UniRef100_Q2J3C8 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Ha... 94 1e-17
UniRef100_Q9SPM1 Extensin-like protein n=1 Tax=Solanum lycopersi... 94 1e-17
UniRef100_Q41707 Extensin class 1 protein n=1 Tax=Vigna unguicul... 94 1e-17
UniRef100_Q39835 Extensin n=1 Tax=Glycine max RepID=Q39835_SOYBN 94 1e-17
UniRef100_B9IAD8 Predicted protein n=1 Tax=Populus trichocarpa R... 94 1e-17
UniRef100_A9P3B7 Anther-specific proline rich protein n=1 Tax=Br... 94 1e-17
UniRef100_A9P3B4 Anther-specific proline rich protein n=1 Tax=Br... 94 1e-17
UniRef100_A9P3B0 Anther-specific proline rich protein n=1 Tax=Br... 94 1e-17
UniRef100_A9P3A9 Anther-specific proline rich protein n=4 Tax=Br... 94 1e-17
UniRef100_A9P3A8 Anther-specific proline rich protein n=1 Tax=Br... 94 1e-17
UniRef100_Q9VAY4 CG5514, isoform A n=1 Tax=Drosophila melanogast... 94 1e-17
UniRef100_Q8MRP6 GH07623p n=1 Tax=Drosophila melanogaster RepID=... 94 1e-17
UniRef100_Q8IMM6 CG5514, isoform B n=1 Tax=Drosophila melanogast... 94 1e-17
UniRef100_B6AGG8 Putative uncharacterized protein n=1 Tax=Crypto... 94 1e-17
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 94 1e-17
UniRef100_Q6CDQ5 YALI0B22110p n=1 Tax=Yarrowia lipolytica RepID=... 94 1e-17
UniRef100_A8NQG1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 94 1e-17
UniRef100_Q03211 Pistil-specific extensin-like protein n=1 Tax=N... 94 1e-17
UniRef100_UPI00005A3746 PREDICTED: similar to Splicing factor 1 ... 93 1e-17
UniRef100_UPI000013DBDC proline-rich protein BstNI subfamily 4 p... 93 1e-17
UniRef100_Q49I31 120 kDa pistil extensin-like protein (Fragment)... 93 1e-17
UniRef100_Q39949 Hydroxyproline-rich protein n=1 Tax=Helianthus ... 93 1e-17
UniRef100_C0PJF1 Putative uncharacterized protein n=1 Tax=Zea ma... 93 1e-17
UniRef100_UPI0000DA2736 PREDICTED: RGD1559532 n=1 Tax=Rattus nor... 93 2e-17
UniRef100_Q04118 Basic salivary proline-rich protein 3 n=1 Tax=H... 93 2e-17
UniRef100_UPI0001984F90 PREDICTED: hypothetical protein n=1 Tax=... 92 2e-17
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 92 2e-17
UniRef100_UPI0000DD9D0C Os11g0657400 n=1 Tax=Oryza sativa Japoni... 92 2e-17
UniRef100_UPI0000221774 Hypothetical protein CBG05727 n=1 Tax=Ca... 92 2e-17
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 92 2e-17
UniRef100_B9G8N7 Putative uncharacterized protein n=1 Tax=Oryza ... 92 2e-17
UniRef100_Q9VVG2 CG13731 n=1 Tax=Drosophila melanogaster RepID=Q... 92 2e-17
UniRef100_A8X1S6 Putative uncharacterized protein n=1 Tax=Caenor... 92 2e-17
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 92 2e-17
UniRef100_A7EJV5 Putative uncharacterized protein n=1 Tax=Sclero... 92 2e-17
UniRef100_UPI00005A198E PREDICTED: similar to cyclin K n=1 Tax=C... 92 3e-17
UniRef100_UPI0000E67202 proline-rich protein BstNI subfamily 3 p... 92 3e-17
UniRef100_Q9XDH2 Proline-rich mucin homolog n=1 Tax=Mycobacteriu... 92 3e-17
UniRef100_Q9LJ64 Extensin protein-like n=1 Tax=Arabidopsis thali... 92 3e-17
UniRef100_Q10MG8 Retrotransposon protein, putative, Ty3-gypsy su... 92 3e-17
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 92 3e-17
UniRef100_B9HRV2 Predicted protein n=1 Tax=Populus trichocarpa R... 92 3e-17
UniRef100_B8LQI7 Putative uncharacterized protein n=1 Tax=Picea ... 92 3e-17
UniRef100_A9NTY4 Putative uncharacterized protein n=1 Tax=Picea ... 92 3e-17
UniRef100_A3AH89 Putative uncharacterized protein n=1 Tax=Oryza ... 92 3e-17
UniRef100_Q2H239 Putative uncharacterized protein n=1 Tax=Chaeto... 92 3e-17
UniRef100_UPI00017F0BD6 PREDICTED: similar to cyclin K n=1 Tax=S... 92 4e-17
UniRef100_UPI00017B0B92 UPI00017B0B92 related cluster n=1 Tax=Te... 92 4e-17
UniRef100_UPI000019B89E proline rich protein 2 (predicted) n=1 T... 92 4e-17
UniRef100_Q04154 Salivary proline-rich protein n=1 Tax=Rattus no... 92 4e-17
UniRef100_Q4PSF3 Proline-rich extensin-like family protein n=1 T... 92 4e-17
UniRef100_O49986 120 kDa style glycoprotein n=1 Tax=Nicotiana al... 92 4e-17
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 92 4e-17
UniRef100_B9RL73 LRX2, putative n=1 Tax=Ricinus communis RepID=B... 92 4e-17
UniRef100_B3M2B3 GF17078 n=1 Tax=Drosophila ananassae RepID=B3M2... 92 4e-17
UniRef100_B0EG62 Inverted formin-2, putative n=1 Tax=Entamoeba d... 92 4e-17
UniRef100_A0EFA7 Chromosome undetermined scaffold_93, whole geno... 92 4e-17
UniRef100_UPI000186EF1C formin 1,2/cappuccino, putative n=1 Tax=... 91 5e-17
UniRef100_UPI000186B03B hypothetical protein BRAFLDRAFT_116781 n... 91 5e-17
UniRef100_UPI0001A2D001 UPI0001A2D001 related cluster n=1 Tax=Da... 91 5e-17
UniRef100_B3DKQ7 Sf1 protein n=1 Tax=Danio rerio RepID=B3DKQ7_DANRE 91 5e-17
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 91 5e-17
UniRef100_B3NWD4 GG19109 (Fragment) n=1 Tax=Drosophila erecta Re... 91 5e-17
UniRef100_B3MVT4 GF22387 n=1 Tax=Drosophila ananassae RepID=B3MV... 91 5e-17
UniRef100_C7YMP5 Putative uncharacterized protein n=1 Tax=Nectri... 91 5e-17
UniRef100_C4XZY9 Putative uncharacterized protein n=1 Tax=Clavis... 91 5e-17
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 91 6e-17
UniRef100_UPI0001A2C288 UPI0001A2C288 related cluster n=1 Tax=Da... 91 6e-17
UniRef100_Q7ZYY2 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 91 6e-17
UniRef100_Q6NYX7 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 91 6e-17
UniRef100_Q5RGR6 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 91 6e-17
UniRef100_Q4RY48 Chromosome 3 SCAF14978, whole genome shotgun se... 91 6e-17
UniRef100_A8WGA1 LOC563976 protein (Fragment) n=1 Tax=Danio reri... 91 6e-17
UniRef100_C1FIW7 Predicted protein n=1 Tax=Micromonas sp. RCC299... 91 6e-17
UniRef100_B4QQM0 GD13257 n=1 Tax=Drosophila simulans RepID=B4QQM... 91 6e-17
UniRef100_UPI000155F323 PREDICTED: splicing factor 1 isoform 2 n... 91 8e-17
UniRef100_UPI0000E22B53 PREDICTED: similar to transcription fact... 91 8e-17
UniRef100_UPI0000E22B51 PREDICTED: splicing factor 1 isoform 14 ... 91 8e-17
UniRef100_UPI00005C06DC PREDICTED: splicing factor 1 isoform 9 n... 91 8e-17
UniRef100_UPI00005A3748 PREDICTED: similar to Splicing factor 1 ... 91 8e-17
UniRef100_UPI00005A3747 PREDICTED: similar to Splicing factor 1 ... 91 8e-17
UniRef100_UPI00001A3E62 PREDICTED: similar to Splicing factor 1 ... 91 8e-17
UniRef100_Q3UI45 Putative uncharacterized protein n=1 Tax=Mus mu... 91 8e-17
UniRef100_B1VUJ2 Putative uncharacterized protein n=1 Tax=Strept... 91 8e-17
UniRef100_Q49I29 120 kDa pistil extensin-like protein (Fragment)... 91 8e-17
UniRef100_B9NAX9 Predicted protein n=1 Tax=Populus trichocarpa R... 91 8e-17
UniRef100_Q86VG2 Splicing factor proline/glutamine-rich (Polypyr... 91 8e-17
UniRef100_Q6PIX2 SFPQ protein (Fragment) n=1 Tax=Homo sapiens Re... 91 8e-17
UniRef100_B4DJU4 cDNA FLJ53344, highly similar to Splicing facto... 91 8e-17
UniRef100_Q2HG22 Putative uncharacterized protein n=1 Tax=Chaeto... 91 8e-17
UniRef100_B6HQ39 Pc22g23920 protein n=1 Tax=Penicillium chrysoge... 91 8e-17
UniRef100_A8NWX4 Putative uncharacterized protein n=1 Tax=Coprin... 91 8e-17
UniRef100_P23246-2 Isoform Short of Splicing factor, proline- an... 91 8e-17
UniRef100_P23246 Splicing factor, proline- and glutamine-rich n=... 91 8e-17
UniRef100_Q64213-3 Isoform 3 of Splicing factor 1 n=1 Tax=Mus mu... 91 8e-17
UniRef100_Q15637 Splicing factor 1 n=3 Tax=Eutheria RepID=SF01_H... 91 8e-17
UniRef100_UPI00015DE8CE diaphanous homolog 1 (Drosophila) n=1 Ta... 90 1e-16
UniRef100_UPI00015DE8CD diaphanous homolog 1 (Drosophila) n=2 Ta... 90 1e-16
UniRef100_UPI0000195290 proline rich protein 2 n=1 Tax=Mus muscu... 90 1e-16
UniRef100_Q5DTQ4 MKIAA4062 protein (Fragment) n=3 Tax=Mus muscul... 90 1e-16
UniRef100_Q09085 Hydroxyproline-rich glycoprotein (HRGP) (Fragme... 90 1e-16
UniRef100_A9P3B9 Anther-specific proline rich protein n=1 Tax=Br... 90 1e-16
UniRef100_Q9VZB2 CG13722 n=1 Tax=Drosophila melanogaster RepID=Q... 90 1e-16
UniRef100_A2DML2 Putative uncharacterized protein n=1 Tax=Tricho... 90 1e-16
UniRef100_B7Z1Q1 cDNA FLJ54737, highly similar to Splicing facto... 90 1e-16
UniRef100_O36027 Wiskott-Aldrich syndrome protein homolog 1 n=1 ... 90 1e-16
UniRef100_O08808 Protein diaphanous homolog 1 n=1 Tax=Mus muscul... 90 1e-16
UniRef100_UPI0001B7BA1F proline-rich protein 15 n=1 Tax=Rattus n... 90 1e-16
UniRef100_Q6NTV3 MGC81512 protein n=1 Tax=Xenopus laevis RepID=Q... 90 1e-16
UniRef100_Q3UK67 Putative uncharacterized protein n=1 Tax=Mus mu... 90 1e-16
UniRef100_C1ECP1 Resistance-nodulation-cell division superfamily... 90 1e-16
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 90 1e-16
UniRef100_Q0ZAL1 Spliceosomal protein on the X n=1 Tax=Bombyx mo... 90 1e-16
UniRef100_B4NUK4 GD24847 n=1 Tax=Drosophila simulans RepID=B4NUK... 90 1e-16
UniRef100_B3LCU8 Putative uncharacterized protein n=1 Tax=Plasmo... 90 1e-16
UniRef100_Q8TF74-2 Isoform 2 of WAS/WASL-interacting protein fam... 90 1e-16
UniRef100_Q8TF74 WAS/WASL-interacting protein family member 2 n=... 90 1e-16
UniRef100_P05143 Proline-rich protein 2 n=1 Tax=Mus musculus Rep... 90 1e-16
UniRef100_UPI00017974A0 PREDICTED: similar to WAS/WASL interacti... 89 2e-16
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 89 2e-16
UniRef100_UPI0001A2BBDD hypothetical protein LOC100009637 n=1 Ta... 89 2e-16
UniRef100_UPI00006A1087 WAS/WASL interacting protein family memb... 89 2e-16
UniRef100_UPI0000250733 proline-rich proteoglycan 2 n=1 Tax=Ratt... 89 2e-16
UniRef100_A2RUY4 Zgc:158395 protein n=1 Tax=Danio rerio RepID=A2... 89 2e-16
UniRef100_Q91X93 Proline-rich protein BstNI subfamily 1 n=1 Tax=... 89 2e-16
UniRef100_A8IEX2 Putative uncharacterized protein n=1 Tax=Azorhi... 89 2e-16
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 89 2e-16
UniRef100_B4PI80 GE20611 n=1 Tax=Drosophila yakuba RepID=B4PI80_... 89 2e-16
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 89 2e-16
UniRef100_A4HCC8 Putative uncharacterized protein n=1 Tax=Leishm... 89 2e-16
UniRef100_Q6CEK4 YALI0B14971p n=1 Tax=Yarrowia lipolytica RepID=... 89 2e-16
UniRef100_Q9Z0G8-2 Isoform 2 of WAS/WASL-interacting protein fam... 89 2e-16
UniRef100_Q9Z0G8 WAS/WASL-interacting protein family member 3 n=... 89 2e-16
UniRef100_P10165 Proline-rich proteoglycan 2 n=1 Tax=Rattus norv... 89 2e-16
UniRef100_Q6PFT9 Amyloid beta A4 precursor protein-binding famil... 89 2e-16
UniRef100_UPI00019859A1 PREDICTED: hypothetical protein n=1 Tax=... 89 2e-16
UniRef100_UPI00019855E0 PREDICTED: hypothetical protein n=1 Tax=... 89 2e-16
UniRef100_UPI0001868875 hypothetical protein BRAFLDRAFT_103538 n... 89 2e-16
UniRef100_UPI0001951234 Ras-associated and pleckstrin homology d... 89 2e-16
UniRef100_Q04117 Salivary proline-rich protein n=1 Tax=Rattus no... 89 2e-16
UniRef100_B4D0M3 Putative uncharacterized protein n=1 Tax=Chthon... 89 2e-16
UniRef100_Q9FSG1 Putative extensin n=1 Tax=Nicotiana sylvestris ... 89 2e-16
UniRef100_Q39864 Hydroxyproline-rich glycoprotein (Fragment) n=1... 89 2e-16
UniRef100_A4QQZ5 Predicted protein n=1 Tax=Magnaporthe grisea Re... 89 2e-16
UniRef100_P05143-2 Isoform 2 of Proline-rich protein 2 n=1 Tax=M... 89 2e-16
UniRef100_UPI0001985C7D PREDICTED: hypothetical protein n=1 Tax=... 89 3e-16
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 89 3e-16
UniRef100_UPI00015DEFCE predicted gene, EG381818 n=1 Tax=Mus mus... 89 3e-16
UniRef100_Q9NZ56 Formin-2 n=2 Tax=Homo sapiens RepID=FMN2_HUMAN 89 3e-16
UniRef100_B8IP63 Putative uncharacterized protein n=1 Tax=Methyl... 89 3e-16
UniRef100_Q9VEP4 CG5225 n=2 Tax=Drosophila melanogaster RepID=Q9... 89 3e-16
UniRef100_B4MN04 GK17614 n=1 Tax=Drosophila willistoni RepID=B4M... 89 3e-16
UniRef100_B4MAS1 GJ15967 n=1 Tax=Drosophila virilis RepID=B4MAS1... 89 3e-16
UniRef100_Q8VIJ6 Splicing factor, proline- and glutamine-rich n=... 89 3e-16
UniRef100_UPI0001A7B0C1 protein binding / structural constituent... 88 4e-16
UniRef100_UPI000155DCD8 PREDICTED: similar to WAS protein family... 88 4e-16
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 88 4e-16
UniRef100_UPI0000E1F800 PREDICTED: similar to SH3-domain interac... 88 4e-16
UniRef100_UPI0000E1F7FD PREDICTED: similar to WASP interacting p... 88 4e-16
UniRef100_UPI00005A14A8 PREDICTED: similar to formin 2 n=1 Tax=C... 88 4e-16
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 88 4e-16
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 88 4e-16
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 88 4e-16
UniRef100_UPI00016E98C2 UPI00016E98C2 related cluster n=1 Tax=Ta... 88 4e-16
UniRef100_UPI0000EB3606 Formin-2. n=1 Tax=Canis lupus familiaris... 88 4e-16
UniRef100_UPI000179F457 PREDICTED: similar to Splicing factor, p... 88 4e-16
UniRef100_UPI0000ECC7C5 enabled homolog n=1 Tax=Gallus gallus Re... 88 4e-16
UniRef100_Q90YB5 AvEna neural variant n=1 Tax=Gallus gallus RepI... 88 4e-16
UniRef100_Q5PPB8 UL36 tegument protein n=2 Tax=Suid herpesvirus ... 88 4e-16
UniRef100_Q4KM71 Splicing factor proline/glutamine rich (Polypyr... 88 4e-16
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 88 4e-16
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 88 4e-16
UniRef100_A8Y2E4 Putative uncharacterized protein (Fragment) n=1... 88 4e-16
UniRef100_A0E1N2 Chromosome undetermined scaffold_73, whole geno... 88 4e-16
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 88 4e-16
UniRef100_Q6IN36 WAS/WASL-interacting protein family member 1 n=... 88 4e-16
UniRef100_P05142 Proline-rich protein HaeIII subfamily 1 n=1 Tax... 88 4e-16
UniRef100_UPI0000F2141A PREDICTED: hypothetical protein n=1 Tax=... 88 5e-16
UniRef100_UPI0000E125D3 Os05g0552600 n=1 Tax=Oryza sativa Japoni... 88 5e-16
UniRef100_UPI0000D9D096 PREDICTED: similar to Wiskott-Aldrich sy... 88 5e-16
UniRef100_UPI0000D99F41 PREDICTED: similar to formin 2 isoform 1... 88 5e-16
UniRef100_UPI0000D99F40 PREDICTED: similar to formin 2 isoform 2... 88 5e-16
UniRef100_UPI0000D8BF7D Amyloid beta A4 precursor protein-bindin... 88 5e-16
UniRef100_UPI00015DEFD1 Proline-rich protein 2 precursor (Prolin... 88 5e-16
UniRef100_UPI0000D61270 WAS/WASL interacting protein family memb... 88 5e-16
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 88 5e-16
UniRef100_A3KD20 Leucine-rich repeat/extensin n=1 Tax=Nicotiana ... 88 5e-16
UniRef100_B4MPC8 GK21628 n=1 Tax=Drosophila willistoni RepID=B4M... 88 5e-16
UniRef100_B8ZZM1 Putative uncharacterized protein WIPF1 n=1 Tax=... 88 5e-16
UniRef100_A3LS13 Predicted protein n=1 Tax=Pichia stipitis RepID... 88 5e-16
UniRef100_Q6PEV3 WAS/WASL-interacting protein family member 2 n=... 88 5e-16
UniRef100_O43516-2 Isoform 2 of WAS/WASL-interacting protein fam... 88 5e-16
UniRef100_O43516 WAS/WASL-interacting protein family member 1 n=... 88 5e-16
UniRef100_UPI00015BB2CD proline rich protein HaeIII subfamily 1 ... 87 7e-16
UniRef100_UPI0000E4896A PREDICTED: similar to CG33556-PA n=1 Tax... 87 7e-16
UniRef100_UPI0000DA1AD6 PREDICTED: hypothetical protein n=1 Tax=... 87 7e-16
UniRef100_Q6PF27 MGC69127 protein n=1 Tax=Xenopus laevis RepID=Q... 87 7e-16
UniRef100_Q9FPQ5 Gamete-specific hydroxyproline-rich glycoprotei... 87 7e-16
UniRef100_Q39353 Cell wall-plasma membrane linker protein n=1 Ta... 87 7e-16
UniRef100_Q54TI7 WH2 domain-containing protein n=1 Tax=Dictyoste... 87 7e-16
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 87 7e-16
UniRef100_C9JDS7 Putative uncharacterized protein ENSP0000039622... 87 7e-16
UniRef100_UPI0001797C5A PREDICTED: similar to formin 2 n=1 Tax=E... 87 9e-16
UniRef100_UPI0000DA3604 PREDICTED: similar to Wire protein n=1 T... 87 9e-16
UniRef100_UPI00004C1216 PREDICTED: similar to WIRE protein n=1 T... 87 9e-16
UniRef100_UPI00001631D5 LRX2 (LEUCINE-RICH REPEAT/EXTENSIN 2); p... 87 9e-16
UniRef100_UPI00015DEFD0 proline rich protein HaeIII subfamily 1 ... 87 9e-16
UniRef100_UPI00016E7DED UPI00016E7DED related cluster n=1 Tax=Ta... 87 9e-16
UniRef100_UPI0000EB3C3B Protein diaphanous homolog 1 (Diaphanous... 87 9e-16
UniRef100_UPI0000EB22A7 WAS/WASL interacting protein family memb... 87 9e-16
UniRef100_UPI0000583117 PREDICTED: similar to WIRE protein isofo... 87 9e-16
UniRef100_Q3KQ40 LOC100101277 protein n=1 Tax=Xenopus laevis Rep... 87 9e-16
UniRef100_Q6X248 UL36 very large tegument protein n=1 Tax=Bovine... 87 9e-16
UniRef100_Q9XIB6 F13F21.7 protein n=1 Tax=Arabidopsis thaliana R... 87 9e-16
UniRef100_Q9LIE9 Similarity to cell wall-plasma membrane linker ... 87 9e-16
UniRef100_O48809 F24O1.18 n=1 Tax=Arabidopsis thaliana RepID=O48... 87 9e-16
UniRef100_Q9GZH1 Putative uncharacterized protein n=1 Tax=Caenor... 87 9e-16
UniRef100_B5DR44 GA28342 n=1 Tax=Drosophila pseudoobscura pseudo... 87 9e-16
UniRef100_B4PI85 GE20610 n=1 Tax=Drosophila yakuba RepID=B4PI85_... 87 9e-16
UniRef100_A2R0A5 Remark: C-terminal truncated ORF due to end of ... 87 9e-16
UniRef100_Q8K1I7 WAS/WASL-interacting protein family member 1 n=... 87 9e-16
UniRef100_Q03173-5 Isoform 4 of Protein enabled homolog n=1 Tax=... 87 9e-16
UniRef100_Q03173-2 Isoform 1 of Protein enabled homolog n=1 Tax=... 87 9e-16
UniRef100_Q03173-4 Isoform 3 of Protein enabled homolog n=1 Tax=... 87 9e-16
UniRef100_Q03173 Protein enabled homolog n=1 Tax=Mus musculus Re... 87 9e-16
UniRef100_Q6NWC6-2 Isoform 2 of Cleavage and polyadenylation spe... 87 9e-16
UniRef100_Q6NWC6 Cleavage and polyadenylation specificity factor... 87 9e-16
UniRef100_UPI0000F2B1F1 PREDICTED: hypothetical protein isoform ... 87 1e-15
UniRef100_UPI000069F62B Formin-2. n=1 Tax=Xenopus (Silurana) tro... 87 1e-15
UniRef100_UPI000021DFA7 PREDICTED: similar to Wire protein n=1 T... 87 1e-15
UniRef100_Q09083 Hydroxyproline-rich glycoprotein n=1 Tax=Phaseo... 87 1e-15
UniRef100_Q01943 Extensin (Class I) (Fragment) n=1 Tax=Solanum l... 87 1e-15
UniRef100_B4PM92 GE24601 n=1 Tax=Drosophila yakuba RepID=B4PM92_... 87 1e-15
UniRef100_B3MUW0 GF21951 n=1 Tax=Drosophila ananassae RepID=B3MU... 87 1e-15
UniRef100_B9ZVX0 Diaphanous homolog 1 (Drosophila), isoform CRA_... 87 1e-15
UniRef100_Q8J1Y5 Wal1 protein n=1 Tax=Eremothecium gossypii RepI... 87 1e-15
UniRef100_Q7RWH7 Cytokinesis protein sepA n=1 Tax=Neurospora cra... 87 1e-15
UniRef100_Q74ZB4 AGR285Wp n=1 Tax=Eremothecium gossypii RepID=Q7... 87 1e-15
UniRef100_O60610-2 Isoform 2 of Protein diaphanous homolog 1 n=1... 87 1e-15
UniRef100_UPI0000DA1DB8 PREDICTED: hypothetical protein n=1 Tax=... 86 2e-15
UniRef100_UPI0000DA1DB7 PREDICTED: hypothetical protein n=1 Tax=... 86 2e-15
UniRef100_UPI00005A5941 PREDICTED: similar to Wiskott-Aldrich sy... 86 2e-15
UniRef100_UPI00006A0E14 Cyclin-K. n=1 Tax=Xenopus (Silurana) tro... 86 2e-15
UniRef100_UPI0001AE73A8 Protein diaphanous homolog 1 (Diaphanous... 86 2e-15
UniRef100_UPI0001AE73A7 Protein diaphanous homolog 1 (Diaphanous... 86 2e-15
UniRef100_UPI000155D622 Protein diaphanous homolog 1 (Diaphanous... 86 2e-15
UniRef100_UPI0000EA87E7 diaphanous 1 isoform 2 n=1 Tax=Homo sapi... 86 2e-15
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 86 2e-15
UniRef100_UPI0000EB01B2 WAS/WASL interacting protein family memb... 86 2e-15
UniRef100_Q5F3Q7 Putative uncharacterized protein n=1 Tax=Gallus... 86 2e-15
UniRef100_Q566H0 MGC115029 protein n=1 Tax=Xenopus laevis RepID=... 86 2e-15
UniRef100_Q0V9U3 Cyclin K n=1 Tax=Xenopus (Silurana) tropicalis ... 86 2e-15
UniRef100_Q49I33 120 kDa pistil extensin-like protein (Fragment)... 86 2e-15
UniRef100_B9GF15 Predicted protein n=1 Tax=Populus trichocarpa R... 86 2e-15
UniRef100_Q18751 Putative uncharacterized protein C50F7.5 n=1 Ta... 86 2e-15
UniRef100_Q6URC4 Diaphanous 1 n=1 Tax=Homo sapiens RepID=Q6URC4_... 86 2e-15
UniRef100_Q17RN4 DIAPH1 protein n=1 Tax=Homo sapiens RepID=Q17RN... 86 2e-15
UniRef100_Q6FSL1 Strain CBS138 chromosome G complete sequence n=... 86 2e-15
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 86 2e-15
UniRef100_A8NEC1 Putative uncharacterized protein n=1 Tax=Coprin... 86 2e-15
UniRef100_O60610 Protein diaphanous homolog 1 n=1 Tax=Homo sapie... 86 2e-15