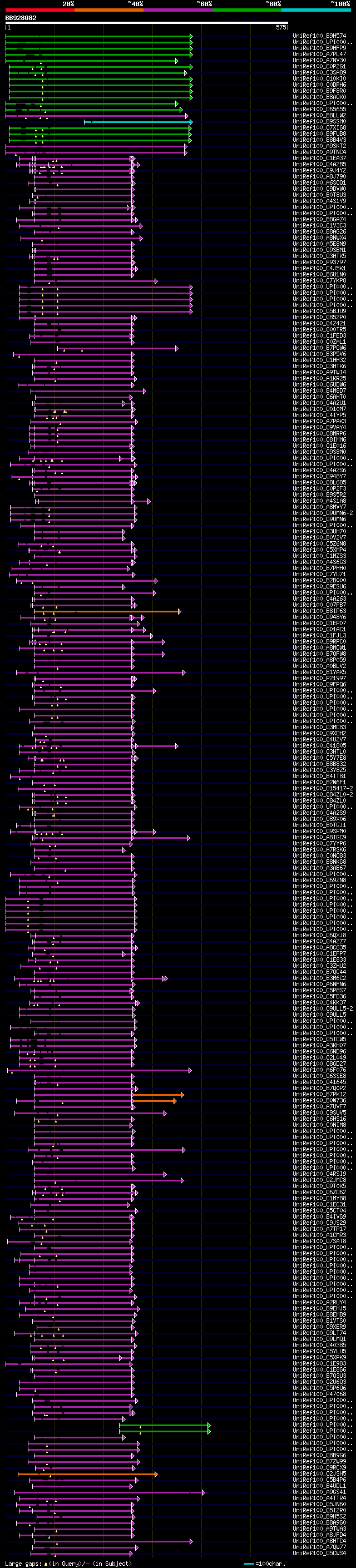
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB928082 RCE39239
(575 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9H574 Predicted protein n=1 Tax=Populus trichocarpa R... 171 4e-41
UniRef100_UPI0001983A58 PREDICTED: hypothetical protein n=1 Tax=... 159 1e-37
UniRef100_B9HFP9 Predicted protein n=1 Tax=Populus trichocarpa R... 159 1e-37
UniRef100_A7PL47 Chromosome chr7 scaffold_20, whole genome shotg... 155 3e-36
UniRef100_A7NV30 Chromosome chr18 scaffold_1, whole genome shotg... 149 2e-34
UniRef100_C0P2G1 Putative uncharacterized protein n=1 Tax=Zea ma... 144 4e-33
UniRef100_C3SA89 SAP domain containing protein n=1 Tax=Brachypod... 132 1e-29
UniRef100_Q10KI0 SAP domain containing protein, expressed n=1 Ta... 129 2e-28
UniRef100_Q0DRH6 Os03g0383800 protein (Fragment) n=1 Tax=Oryza s... 129 2e-28
UniRef100_B9F8R0 Putative uncharacterized protein n=1 Tax=Oryza ... 129 2e-28
UniRef100_B8AQK0 Putative uncharacterized protein n=1 Tax=Oryza ... 127 5e-28
UniRef100_UPI0001985479 PREDICTED: hypothetical protein n=1 Tax=... 127 8e-28
UniRef100_O65655 Putative uncharacterized protein AT4g39680 n=1 ... 127 8e-28
UniRef100_B8LLW2 Putative uncharacterized protein n=1 Tax=Picea ... 120 6e-26
UniRef100_B9SSM0 Putative uncharacterized protein n=1 Tax=Ricinu... 119 1e-25
UniRef100_Q7XIG8 Os07g0626200 protein n=1 Tax=Oryza sativa Japon... 112 2e-23
UniRef100_B9FUB8 Putative uncharacterized protein n=1 Tax=Oryza ... 112 2e-23
UniRef100_B8B4V3 Putative uncharacterized protein n=1 Tax=Oryza ... 112 2e-23
UniRef100_A9SKT2 Predicted protein n=1 Tax=Physcomitrella patens... 78 6e-13
UniRef100_A9TNC4 Predicted protein n=1 Tax=Physcomitrella patens... 66 2e-09
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 66 2e-09
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 64 6e-09
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 64 1e-08
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 63 1e-08
UniRef100_A6SQQ1 Putative uncharacterized protein n=1 Tax=Botryo... 63 1e-08
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 63 2e-08
UniRef100_B0T8U3 Peptidase M56 BlaR1 n=1 Tax=Caulobacter sp. K31... 63 2e-08
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 62 3e-08
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 62 4e-08
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_B8GAZ4 Putative uncharacterized protein n=1 Tax=Chloro... 61 5e-08
UniRef100_C1V3C3 TonB family protein n=1 Tax=Haliangium ochraceu... 61 5e-08
UniRef100_B8AG26 Putative uncharacterized protein n=1 Tax=Oryza ... 61 5e-08
UniRef100_A8NWX4 Putative uncharacterized protein n=1 Tax=Coprin... 61 5e-08
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 61 7e-08
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 61 7e-08
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 61 7e-08
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 61 7e-08
UniRef100_C4J5K1 Putative uncharacterized protein n=1 Tax=Zea ma... 60 9e-08
UniRef100_B6U1N0 Putative uncharacterized protein n=1 Tax=Zea ma... 60 9e-08
UniRef100_C7YKP8 Predicted protein n=1 Tax=Nectria haematococca ... 60 9e-08
UniRef100_UPI0000DA3CE1 PREDICTED: similar to Apoptotic chromati... 60 1e-07
UniRef100_UPI0001B7A155 UPI0001B7A155 related cluster n=1 Tax=Ra... 60 1e-07
UniRef100_UPI0000501C1A UPI0000501C1A related cluster n=1 Tax=Ra... 60 1e-07
UniRef100_UPI0000250AA8 PREDICTED: similar to Apoptotic chromati... 60 1e-07
UniRef100_Q5BJU9 Acin1 protein (Fragment) n=1 Tax=Rattus norvegi... 60 1e-07
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 60 1e-07
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 60 1e-07
UniRef100_Q00TR5 Homology to unknown gene n=1 Tax=Ostreococcus t... 60 1e-07
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 1e-07
UniRef100_Q0ZAL1 Spliceosomal protein on the X n=1 Tax=Bombyx mo... 60 1e-07
UniRef100_B7PGW6 Acinus, putative (Fragment) n=1 Tax=Ixodes scap... 60 1e-07
UniRef100_B3P5V6 GG11582 n=1 Tax=Drosophila erecta RepID=B3P5V6_... 60 1e-07
UniRef100_Q1HH32 Putative uncharacterized protein n=1 Tax=Anther... 60 2e-07
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 60 2e-07
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 60 2e-07
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 60 2e-07
UniRef100_Q6UDW6 Erythrocyte membrane protein 1 n=1 Tax=Plasmodi... 60 2e-07
UniRef100_B4M8D7 GJ16621 n=1 Tax=Drosophila virilis RepID=B4M8D7... 60 2e-07
UniRef100_Q6AHT0 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 60 2e-07
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 59 2e-07
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 59 2e-07
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 59 2e-07
UniRef100_A7PAK3 Chromosome chr14 scaffold_9, whole genome shotg... 59 2e-07
UniRef100_Q9VAY4 CG5514, isoform A n=1 Tax=Drosophila melanogast... 59 2e-07
UniRef100_Q8MRP6 GH07623p n=1 Tax=Drosophila melanogaster RepID=... 59 2e-07
UniRef100_Q8IMM6 CG5514, isoform B n=1 Tax=Drosophila melanogast... 59 2e-07
UniRef100_Q1E016 Predicted protein n=1 Tax=Coccidioides immitis ... 59 2e-07
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 59 2e-07
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 59 3e-07
UniRef100_UPI0000E25181 PREDICTED: myeloid/lymphoid or mixed-lin... 59 3e-07
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 59 3e-07
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 59 3e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 59 3e-07
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 59 3e-07
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 59 3e-07
UniRef100_A4S1A8 Predicted protein n=1 Tax=Ostreococcus lucimari... 59 3e-07
UniRef100_A8MVY7 Putative uncharacterized protein ENSP0000038013... 59 3e-07
UniRef100_Q9UMN6-2 Isoform 2 of Histone-lysine N-methyltransfera... 59 3e-07
UniRef100_Q9UMN6 Histone-lysine N-methyltransferase MLL4 n=1 Tax... 59 3e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 59 3e-07
UniRef100_Q3UH70 Putative uncharacterized protein n=1 Tax=Mus mu... 59 3e-07
UniRef100_B0V2V7 Bromodomain containing 4 n=1 Tax=Mus musculus R... 59 3e-07
UniRef100_C5Z6N8 Putative uncharacterized protein Sb10g025205 (F... 59 3e-07
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 59 3e-07
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 3e-07
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 59 3e-07
UniRef100_B7PHH0 Cyclin t, putative n=1 Tax=Ixodes scapularis Re... 59 3e-07
UniRef100_C7YU71 Predicted protein n=1 Tax=Nectria haematococca ... 59 3e-07
UniRef100_B2B000 Predicted CDS Pa_3_2220 n=1 Tax=Podospora anser... 59 3e-07
UniRef100_Q9ESU6 Bromodomain-containing protein 4 n=1 Tax=Mus mu... 59 3e-07
UniRef100_UPI00017F00CB PREDICTED: similar to protocadherin 15 n... 58 5e-07
UniRef100_Q4A263 Putative membrane protein n=1 Tax=Emiliania hux... 58 5e-07
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 58 5e-07
UniRef100_B8IP63 Putative uncharacterized protein n=1 Tax=Methyl... 58 5e-07
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 58 5e-07
UniRef100_Q1EP07 Putative uncharacterized protein n=1 Tax=Musa a... 58 5e-07
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 58 5e-07
UniRef100_C1FJL3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 58 5e-07
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 58 5e-07
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 58 5e-07
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 58 5e-07
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 58 5e-07
UniRef100_A0BLV2 Chromosome undetermined scaffold_115, whole gen... 58 5e-07
UniRef100_B1YAK5 Putative uncharacterized protein n=1 Tax=Thermo... 58 5e-07
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 58 5e-07
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 58 5e-07
UniRef100_UPI0001868875 hypothetical protein BRAFLDRAFT_103538 n... 58 6e-07
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 58 6e-07
UniRef100_UPI00005A14A8 PREDICTED: similar to formin 2 n=1 Tax=C... 58 6e-07
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 58 6e-07
UniRef100_UPI0000EB3606 Formin-2. n=1 Tax=Canis lupus familiaris... 58 6e-07
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 58 6e-07
UniRef100_Q3MC83 Putative uncharacterized protein n=1 Tax=Anabae... 58 6e-07
UniRef100_Q9XDH2 Proline-rich mucin homolog n=1 Tax=Mycobacteriu... 58 6e-07
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 58 6e-07
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 58 6e-07
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 58 6e-07
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 58 6e-07
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 58 6e-07
UniRef100_C3Y8Z5 Putative uncharacterized protein n=1 Tax=Branch... 58 6e-07
UniRef100_B4IT81 GE19010 n=1 Tax=Drosophila yakuba RepID=B4IT81_... 58 6e-07
UniRef100_B2W6F1 Putative uncharacterized protein n=1 Tax=Pyreno... 58 6e-07
UniRef100_O15417-2 Isoform 2 of Trinucleotide repeat-containing ... 58 6e-07
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 58 6e-07
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 58 6e-07
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 57 8e-07
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 57 8e-07
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 57 8e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 57 8e-07
UniRef100_Q9SPM0 Extensin-like protein n=1 Tax=Zea mays RepID=Q9... 57 8e-07
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 57 8e-07
UniRef100_Q7YYP6 Hydroxyproline-rich glycoprotein dz-hrgp, proba... 57 8e-07
UniRef100_A7RSK6 Predicted protein (Fragment) n=1 Tax=Nematostel... 57 8e-07
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 57 8e-07
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 57 8e-07
UniRef100_A3AB67 Formin-like protein 16 n=1 Tax=Oryza sativa Jap... 57 8e-07
UniRef100_UPI0000D9EBB1 PREDICTED: myeloid/lymphoid or mixed-lin... 57 1e-06
UniRef100_Q69ZN8 MKIAA1205 protein (Fragment) n=3 Tax=Mus muscul... 57 1e-06
UniRef100_UPI00015DF9DB Proline-rich protein 12. n=1 Tax=Homo sa... 57 1e-06
UniRef100_UPI0001596889 proline rich 12 n=1 Tax=Homo sapiens Rep... 57 1e-06
UniRef100_UPI00016E59B7 UPI00016E59B7 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E59B6 UPI00016E59B6 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E599B UPI00016E599B related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E599A UPI00016E599A related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E5999 UPI00016E5999 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E595C UPI00016E595C related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_Q6QXJ8 ORF55 n=1 Tax=Agrotis segetum granulovirus RepI... 57 1e-06
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 57 1e-06
UniRef100_A8C635 Putative uncharacterized protein n=1 Tax=Anther... 57 1e-06
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 57 1e-06
UniRef100_C1E833 Predicted protein n=1 Tax=Micromonas sp. RCC299... 57 1e-06
UniRef100_C3ZHU2 Putative uncharacterized protein n=1 Tax=Branch... 57 1e-06
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 57 1e-06
UniRef100_B3M6C2 GF24314 n=1 Tax=Drosophila ananassae RepID=B3M6... 57 1e-06
UniRef100_A6NFN6 Putative uncharacterized protein PRR12 n=1 Tax=... 57 1e-06
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 57 1e-06
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 57 1e-06
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 57 1e-06
UniRef100_Q9ULL5-2 Isoform 2 of Proline-rich protein 12 n=1 Tax=... 57 1e-06
UniRef100_Q9ULL5 Proline-rich protein 12 n=1 Tax=Homo sapiens Re... 57 1e-06
UniRef100_UPI000023E328 hypothetical protein FG01070.1 n=1 Tax=G... 57 1e-06
UniRef100_UPI0001A2C549 protocadherin 15b n=1 Tax=Danio rerio Re... 57 1e-06
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_Q5ICW5 Protocadherin 15b n=1 Tax=Danio rerio RepID=Q5I... 57 1e-06
UniRef100_A3KH07 Protocadherin 15b (Fragment) n=1 Tax=Danio reri... 57 1e-06
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 57 1e-06
UniRef100_Q2L049 Filamentous hemagglutinin/adhesin n=1 Tax=Borde... 57 1e-06
UniRef100_Q8GD27 Adhesin FhaB n=1 Tax=Bordetella avium RepID=Q8G... 57 1e-06
UniRef100_A6F076 Putative uncharacterized protein n=1 Tax=Marino... 57 1e-06
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 57 1e-06
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 57 1e-06
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_B0W736 Formin 1,2/cappuccino n=1 Tax=Culex quinquefasc... 57 1e-06
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 57 1e-06
UniRef100_C9SUV5 Predicted protein n=1 Tax=Verticillium albo-atr... 57 1e-06
UniRef100_C6HS16 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 57 1e-06
UniRef100_C0NIM8 Putative uncharacterized protein n=1 Tax=Ajello... 57 1e-06
UniRef100_UPI00017C2B21 PREDICTED: similar to WAS/WASL interacti... 56 2e-06
UniRef100_UPI0001795EB6 PREDICTED: similar to WAS/WASL interacti... 56 2e-06
UniRef100_UPI0001792D8C PREDICTED: similar to formin 3 CG33556-P... 56 2e-06
UniRef100_UPI0000E4A5ED PREDICTED: similar to CCDC33 protein n=2... 56 2e-06
UniRef100_UPI0000E1F0D2 PREDICTED: formin 2 n=1 Tax=Pan troglody... 56 2e-06
UniRef100_UPI00016E0FDB UPI00016E0FDB related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_UPI000179ED76 UPI000179ED76 related cluster n=1 Tax=Bo... 56 2e-06
UniRef100_Q4RSI9 Chromosome 13 SCAF15000, whole genome shotgun s... 56 2e-06
UniRef100_Q2JMC8 TonB family protein n=1 Tax=Synechococcus sp. J... 56 2e-06
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 56 2e-06
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 56 2e-06
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 56 2e-06
UniRef100_C1EC31 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 2e-06
UniRef100_Q5CT04 Putative uncharacterized protein n=1 Tax=Crypto... 56 2e-06
UniRef100_B4IVG9 GE14936 n=1 Tax=Drosophila yakuba RepID=B4IVG9_... 56 2e-06
UniRef100_C9JS29 Putative uncharacterized protein TNRC18 n=1 Tax... 56 2e-06
UniRef100_A7TP17 Putative uncharacterized protein n=1 Tax=Vander... 56 2e-06
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 56 2e-06
UniRef100_Q7SAT8 Actin cytoskeleton-regulatory complex protein p... 56 2e-06
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI000186476A hypothetical protein BRAFLDRAFT_84276 n=... 56 2e-06
UniRef100_UPI0000DB7674 PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000DA1CDB PREDICTED: similar to CG17233-PA, isofor... 56 2e-06
UniRef100_UPI0000DA19B0 PREDICTED: similar to CG17233-PA, isofor... 56 2e-06
UniRef100_UPI0000D9ED26 PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0001A2BBDD hypothetical protein LOC100009637 n=1 Ta... 56 2e-06
UniRef100_UPI0001B7BF44 UPI0001B7BF44 related cluster n=1 Tax=Ra... 56 2e-06
UniRef100_UPI00016E597F UPI00016E597F related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_A2RUY4 Zgc:158395 protein n=1 Tax=Danio rerio RepID=A2... 56 2e-06
UniRef100_B9EHJ5 Fmn1 protein n=1 Tax=Mus musculus RepID=B9EHJ5_... 56 2e-06
UniRef100_B8EMB9 TonB family protein n=1 Tax=Methylocella silves... 56 2e-06
UniRef100_B1VTS0 Putative uncharacterized protein n=1 Tax=Strept... 56 2e-06
UniRef100_Q9XER9 Putative transcription factor n=1 Tax=Arabidops... 56 2e-06
UniRef100_Q9LT74 Similarity to late embryogenesis abundant prote... 56 2e-06
UniRef100_Q9LMQ1 F7H2.17 protein n=1 Tax=Arabidopsis thaliana Re... 56 2e-06
UniRef100_Q40385 Pistil extensin-like protein n=1 Tax=Nicotiana ... 56 2e-06
UniRef100_C5YLU5 Putative uncharacterized protein Sb07g000890 n=... 56 2e-06
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 56 2e-06
UniRef100_C1E983 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 2e-06
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 2e-06
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 56 2e-06
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 56 2e-06
UniRef100_C5P6Q6 WH1 domain containing protein n=1 Tax=Coccidioi... 56 2e-06
UniRef100_P47068 Myosin tail region-interacting protein MTI1 n=1... 56 2e-06
UniRef100_UPI0001982E0B PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0001797605 PREDICTED: bromodomain containing 4 n=1 ... 55 3e-06
UniRef100_UPI0000E4764B PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0000DA2C5A bromodomain containing 4 n=1 Tax=Rattus ... 55 3e-06
UniRef100_UPI00017B4375 UPI00017B4375 related cluster n=1 Tax=Te... 55 3e-06
UniRef100_UPI00017B4374 UPI00017B4374 related cluster n=1 Tax=Te... 55 3e-06
UniRef100_UPI0000DBF793 Brd4 protein n=1 Tax=Rattus norvegicus R... 55 3e-06
UniRef100_UPI0000566A27 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 55 3e-06
UniRef100_UPI0000566A26 formin 1 n=1 Tax=Mus musculus RepID=UPI0... 55 3e-06
UniRef100_Q8B9G6 Putative uncharacterized protein n=1 Tax=Rachip... 55 3e-06
UniRef100_B7ZW99 Fmn1 protein n=1 Tax=Mus musculus RepID=B7ZW99_... 55 3e-06
UniRef100_Q9RCX9 Putative secreted proline-rich protein n=1 Tax=... 55 3e-06
UniRef100_Q2JSH5 Protein kinase n=1 Tax=Synechococcus sp. JA-3-3... 55 3e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 55 3e-06
UniRef100_B4UDL1 Putative uncharacterized protein n=1 Tax=Anaero... 55 3e-06
UniRef100_A9GS41 Putative uncharacterized protein n=1 Tax=Sorang... 55 3e-06
UniRef100_A4TTR4 Putative uncharacterized protein n=1 Tax=Magnet... 55 3e-06
UniRef100_Q5JN60 Os01g0750600 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 55 3e-06
UniRef100_B9H5S2 Predicted protein n=1 Tax=Populus trichocarpa R... 55 3e-06
UniRef100_B8A9G0 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 55 3e-06
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 55 3e-06
UniRef100_A8HTC4 Predicted protein n=1 Tax=Chlamydomonas reinhar... 55 3e-06
UniRef100_A7QW77 Chromosome chr3 scaffold_199, whole genome shot... 55 3e-06
UniRef100_Q5CWS4 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_Q5CLM1 Putative uncharacterized protein n=1 Tax=Crypto... 55 3e-06
UniRef100_A0E1N2 Chromosome undetermined scaffold_73, whole geno... 55 3e-06
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 55 3e-06
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 55 3e-06
UniRef100_B8PC49 Predicted protein n=1 Tax=Postia placenta Mad-6... 55 3e-06
UniRef100_B2VSJ8 Putative uncharacterized protein n=1 Tax=Pyreno... 55 3e-06
UniRef100_B2AUP9 Predicted CDS Pa_1_19860 n=1 Tax=Podospora anse... 55 3e-06
UniRef100_A8N8M7 Putative uncharacterized protein n=1 Tax=Coprin... 55 3e-06
UniRef100_Q05860-5 Isoform 5 of Formin-1 n=1 Tax=Mus musculus Re... 55 3e-06
UniRef100_Q05860-2 Isoform 2 of Formin-1 n=1 Tax=Mus musculus Re... 55 3e-06
UniRef100_Q05860-3 Isoform 3 of Formin-1 n=1 Tax=Mus musculus Re... 55 3e-06
UniRef100_Q05860-6 Isoform 6 of Formin-1 n=1 Tax=Mus musculus Re... 55 3e-06
UniRef100_Q05860 Formin-1 n=1 Tax=Mus musculus RepID=FMN1_MOUSE 55 3e-06
UniRef100_UPI0001A7B186 actin binding n=1 Tax=Arabidopsis thalia... 51 4e-06
UniRef100_Q9LVN1 Formin-like protein 13 n=1 Tax=Arabidopsis thal... 51 4e-06
UniRef100_UPI0001984CA4 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0001983015 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0000E47360 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0000DD9D0C Os11g0657400 n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_B0T1Q0 Peptidase M23B n=1 Tax=Caulobacter sp. K31 RepI... 55 4e-06
UniRef100_A0LDK3 TonB family protein n=1 Tax=Magnetococcus sp. M... 55 4e-06
UniRef100_C9KBI4 Cell wall-associated hydrolase, invasion-associ... 55 4e-06
UniRef100_B9NPS1 Possible TolA protein n=1 Tax=Rhodobacteraceae ... 55 4e-06
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 55 4e-06
UniRef100_C0Z219 AT2G14890 protein n=1 Tax=Arabidopsis thaliana ... 55 4e-06
UniRef100_C0PPC0 Putative uncharacterized protein n=1 Tax=Zea ma... 55 4e-06
UniRef100_B9G8N7 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 55 4e-06
UniRef100_C3Z6S3 Putative uncharacterized protein n=1 Tax=Branch... 55 4e-06
UniRef100_B4NYZ4 GE18300 n=1 Tax=Drosophila yakuba RepID=B4NYZ4_... 55 4e-06
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 55 4e-06
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 55 4e-06
UniRef100_Q4WCV2 Actin associated protein Wsp1, putative n=1 Tax... 55 4e-06
UniRef100_C4JK96 Putative uncharacterized protein n=1 Tax=Uncino... 55 4e-06
UniRef100_B8M4W4 Actin associated protein Wsp1, putative n=1 Tax... 55 4e-06
UniRef100_B8M4W3 Actin associated protein Wsp1, putative n=1 Tax... 55 4e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 55 4e-06
UniRef100_Q91Z58 Uncharacterized protein C6orf132 homolog n=1 Ta... 55 4e-06
UniRef100_O60885 Bromodomain-containing protein 4 n=1 Tax=Homo s... 55 4e-06
UniRef100_UPI0001982E1E PREDICTED: hypothetical protein n=1 Tax=... 55 5e-06
UniRef100_UPI0001795912 PREDICTED: similar to protocadherin 15 n... 55 5e-06
UniRef100_UPI0001552DAA PREDICTED: similar to CR16 n=1 Tax=Mus m... 55 5e-06
UniRef100_UPI0000E49EF3 PREDICTED: hypothetical protein n=1 Tax=... 55 5e-06
UniRef100_UPI0000E481CB PREDICTED: hypothetical protein n=1 Tax=... 55 5e-06
UniRef100_UPI00005A499E PREDICTED: similar to CG40351-PA.3 n=1 T... 55 5e-06
UniRef100_UPI000184A044 SET domain-containing protein 1B n=1 Tax... 55 5e-06
UniRef100_Q1JQ01 Acin1a protein n=1 Tax=Danio rerio RepID=Q1JQ01... 55 5e-06
UniRef100_B0R0H3 Novel protein similar to human apoptotic chroma... 55 5e-06
UniRef100_Q8QLN2 Ac1629 n=1 Tax=Mamestra configurata NPV-A RepID... 55 5e-06
UniRef100_Q0RSN4 Putative uncharacterized protein n=1 Tax=Franki... 55 5e-06
UniRef100_Q0BT49 Hypothetical secreted protein n=1 Tax=Granuliba... 55 5e-06
UniRef100_A9F6B6 Putative uncharacterized protein n=1 Tax=Sorang... 55 5e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 55 5e-06
UniRef100_A1TSD3 TonB family protein n=1 Tax=Acidovorax citrulli... 55 5e-06
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 55 5e-06
UniRef100_Q4U2V9 Hydroxyproline-rich glycoprotein GAS30 n=1 Tax=... 55 5e-06
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 55 5e-06
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 55 5e-06
UniRef100_B9RQE5 Putative uncharacterized protein n=1 Tax=Ricinu... 55 5e-06
UniRef100_B4FZM6 Putative uncharacterized protein n=1 Tax=Zea ma... 55 5e-06
UniRef100_A9RE34 Predicted protein n=1 Tax=Physcomitrella patens... 55 5e-06
UniRef100_A8JBZ3 Metalloproteinase of VMP family n=1 Tax=Chlamyd... 55 5e-06
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 55 5e-06
UniRef100_A5BR16 Putative uncharacterized protein n=1 Tax=Vitis ... 55 5e-06
UniRef100_A9V3V4 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 5e-06
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 55 5e-06
UniRef100_C5G899 Predicted protein n=1 Tax=Ajellomyces dermatiti... 55 5e-06
UniRef100_C4JT61 Predicted protein n=1 Tax=Uncinocarpus reesii 1... 55 5e-06
UniRef100_C0NGH8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 55 5e-06
UniRef100_P0C7L0-2 Isoform 2 of WAS/WASL-interacting protein fam... 55 5e-06
UniRef100_P0C7L0 WAS/WASL-interacting protein family member 3 n=... 55 5e-06
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 55 5e-06
UniRef100_UPI000198409E PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI000198296E PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI00018298BF putative membrane protein n=1 Tax=Emilia... 54 7e-06
UniRef100_UPI0001797C5A PREDICTED: similar to formin 2 n=1 Tax=E... 54 7e-06
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 54 7e-06
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0000DA1981 PREDICTED: similar to WW domain binding ... 54 7e-06
UniRef100_UPI0000D9EA10 PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0000DC200F UPI0000DC200F related cluster n=1 Tax=Ra... 54 7e-06
UniRef100_UPI00015AA540 WW domain binding protein 7 n=1 Tax=Mus ... 54 7e-06
UniRef100_UPI0000511CC2 WW domain binding protein 7 n=1 Tax=Mus ... 54 7e-06
UniRef100_Q4A2U2 Putative membrane protein n=1 Tax=Emiliania hux... 54 7e-06
UniRef100_Q825Z2 Putative proline-rich protein n=1 Tax=Streptomy... 54 7e-06
UniRef100_A9HB01 Putative uncharacterized protein n=1 Tax=Glucon... 54 7e-06
UniRef100_Q7P3A4 Hypothetical WASP N-WASP MENA proteins n=1 Tax=... 54 7e-06
UniRef100_C9Z517 Putative secreted proline-rich protein n=1 Tax=... 54 7e-06
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 54 7e-06
UniRef100_Q49I32 120 kDa pistil extensin-like protein (Fragment)... 54 7e-06
UniRef100_Q49I27 120 kDa pistil extensin-like protein (Fragment)... 54 7e-06
UniRef100_Q40549 Pistil extensin like protein, partial CDS (Frag... 54 7e-06
UniRef100_Q39353 Cell wall-plasma membrane linker protein n=1 Ta... 54 7e-06
UniRef100_Q2QWV3 Transposon protein, putative, CACTA, En/Spm sub... 54 7e-06
UniRef100_C5YLB8 Putative uncharacterized protein Sb07g000099 n=... 54 7e-06
UniRef100_C5YBI1 Putative uncharacterized protein Sb06g021550 n=... 54 7e-06
UniRef100_B9S0D2 Splicing factor 3A subunit, putative n=1 Tax=Ri... 54 7e-06
UniRef100_B9RFR5 Mads box protein, putative n=1 Tax=Ricinus comm... 54 7e-06
UniRef100_B9HGL7 Predicted protein n=1 Tax=Populus trichocarpa R... 54 7e-06
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 54 7e-06
UniRef100_A7P2V2 Chromosome chr1 scaffold_5, whole genome shotgu... 54 7e-06
UniRef100_A5C8X5 Putative uncharacterized protein n=1 Tax=Vitis ... 54 7e-06
UniRef100_A2XD25 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_Q9VJ12 Hook-like, isoform A n=1 Tax=Drosophila melanog... 54 7e-06
UniRef100_Q95UQ2 Subtilisin-like protein n=1 Tax=Toxoplasma gond... 54 7e-06
UniRef100_Q581D1 Flagellum-adhesion glycoprotein, putative n=1 T... 54 7e-06
UniRef100_Q581C6 Flagellum-adhesion glycoprotein, putative n=1 T... 54 7e-06
UniRef100_B9PU53 Subtilase family protein n=1 Tax=Toxoplasma gon... 54 7e-06
UniRef100_B4IVK7 GE14960 n=1 Tax=Drosophila yakuba RepID=B4IVK7_... 54 7e-06
UniRef100_B4IT39 GE18257 n=1 Tax=Drosophila yakuba RepID=B4IT39_... 54 7e-06
UniRef100_B4IGY1 GM16364 n=1 Tax=Drosophila sechellia RepID=B4IG... 54 7e-06
UniRef100_A7SGL4 Predicted protein n=1 Tax=Nematostella vectensi... 54 7e-06
UniRef100_A4H939 Formin, putative n=1 Tax=Leishmania braziliensi... 54 7e-06
UniRef100_Q96VI4 Protease 1 n=1 Tax=Pneumocystis carinii RepID=Q... 54 7e-06
UniRef100_Q5KGJ5 Putative uncharacterized protein n=1 Tax=Filoba... 54 7e-06
UniRef100_C5JPY4 Putative uncharacterized protein n=1 Tax=Ajello... 54 7e-06
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 54 7e-06
UniRef100_O08550 Histone-lysine N-methyltransferase MLL4 n=1 Tax... 54 7e-06
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI00019259C5 PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 54 9e-06
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 54 9e-06
UniRef100_UPI0000D9E432 PREDICTED: similar to formin-like 1 isof... 54 9e-06
UniRef100_UPI0000D9E42C PREDICTED: similar to formin-like 1 isof... 54 9e-06
UniRef100_UPI0001B7A7A5 UPI0001B7A7A5 related cluster n=1 Tax=Ra... 54 9e-06
UniRef100_Q070M8 Putative uncharacterized protein n=1 Tax=Crocod... 54 9e-06
UniRef100_B7FEG6 Very large tegument protein n=1 Tax=Equid herpe... 54 9e-06
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 54 9e-06
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 54 9e-06
UniRef100_B9LBU3 Putative uncharacterized protein n=1 Tax=Chloro... 54 9e-06
UniRef100_A9WHI6 Putative uncharacterized protein n=1 Tax=Chloro... 54 9e-06
UniRef100_A4TBI8 Putative uncharacterized protein n=1 Tax=Mycoba... 54 9e-06
UniRef100_Q6YWB1 Putative uncharacterized protein P0501E09.22 n=... 54 9e-06
UniRef100_Q013M1 Chromosome 08 contig 1, DNA sequence n=1 Tax=Os... 54 9e-06
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 54 9e-06
UniRef100_C5XS10 Putative uncharacterized protein Sb04g033190 n=... 54 9e-06
UniRef100_C5XK46 Putative uncharacterized protein Sb03g034710 n=... 54 9e-06
UniRef100_C1FJL2 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 9e-06
UniRef100_B9IJ95 Predicted protein n=1 Tax=Populus trichocarpa R... 54 9e-06
UniRef100_B9FY87 Putative uncharacterized protein n=1 Tax=Oryza ... 54 9e-06
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 54 9e-06
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 54 9e-06
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 54 9e-06
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 54 9e-06
UniRef100_Q29N97 GA10336 n=1 Tax=Drosophila pseudoobscura pseudo... 54 9e-06
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 54 9e-06
UniRef100_B4Q9F1 GD24176 n=1 Tax=Drosophila simulans RepID=B4Q9F... 54 9e-06
UniRef100_B4PAF7 GE13225 n=1 Tax=Drosophila yakuba RepID=B4PAF7_... 54 9e-06
UniRef100_B4JDP7 Expanded n=1 Tax=Drosophila grimshawi RepID=B4J... 54 9e-06
UniRef100_B4I5Q2 GM17317 n=1 Tax=Drosophila sechellia RepID=B4I5... 54 9e-06
UniRef100_B4G826 GL18912 n=1 Tax=Drosophila persimilis RepID=B4G... 54 9e-06
UniRef100_B3NMA5 GG21152 n=1 Tax=Drosophila erecta RepID=B3NMA5_... 54 9e-06
UniRef100_B3M929 GF25073 n=1 Tax=Drosophila ananassae RepID=B3M9... 54 9e-06
UniRef100_A0E3T6 Chromosome undetermined scaffold_77, whole geno... 54 9e-06
UniRef100_Q9UVD0 Kexin-like serine endoprotease (Fragment) n=1 T... 54 9e-06
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 54 9e-06
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 54 9e-06
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 54 9e-06
UniRef100_B6H7L0 Pc16g07390 protein n=1 Tax=Penicillium chrysoge... 54 9e-06