[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB925032 RCE33796
(548 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
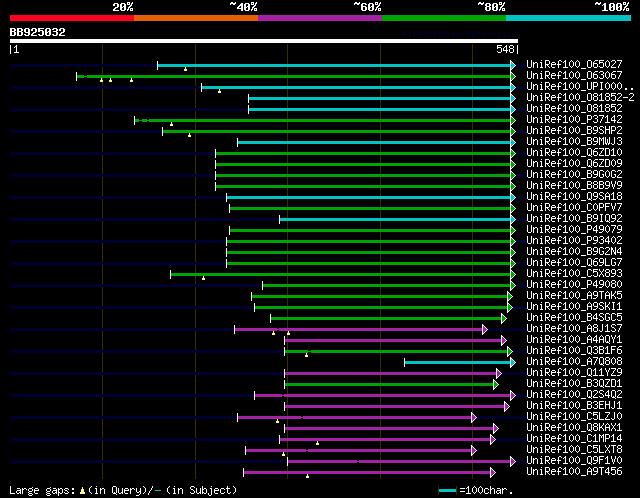
significant alignments:[graphical|details]
UniRef100_O65027 Aspartokinase-homoserine dehydrogenase (Fragmen... 177 3e-43
UniRef100_O63067 Aspartokinase-homoserine dehydrogenase n=1 Tax=... 172 1e-41
UniRef100_UPI00019855EA PREDICTED: hypothetical protein n=1 Tax=... 152 2e-35
UniRef100_O81852-2 Isoform 2 of Bifunctional aspartokinase/homos... 150 8e-35
UniRef100_O81852 Homoserine dehydrogenase n=2 Tax=Arabidopsis th... 150 8e-35
UniRef100_P37142 Homoserine dehydrogenase (Fragment) n=1 Tax=Dau... 147 6e-34
UniRef100_B9SHP2 Aspartate kinase, putative n=1 Tax=Ricinus comm... 144 3e-33
UniRef100_B9MWJ3 Predicted protein (Fragment) n=1 Tax=Populus tr... 143 7e-33
UniRef100_Q6ZD10 Putative aspartate kinase, homoserine dehydroge... 139 1e-31
UniRef100_Q6ZD09 Putative aspartate kinase, homoserine dehydroge... 139 1e-31
UniRef100_B9G0G2 Putative uncharacterized protein n=1 Tax=Oryza ... 139 1e-31
UniRef100_B8B9V9 Putative uncharacterized protein n=1 Tax=Oryza ... 139 1e-31
UniRef100_Q9SA18 Homoserine dehydrogenase n=1 Tax=Arabidopsis th... 138 3e-31
UniRef100_C0PFV7 Putative uncharacterized protein n=1 Tax=Zea ma... 137 7e-31
UniRef100_B9IQ92 Predicted protein n=1 Tax=Populus trichocarpa R... 135 1e-30
UniRef100_P49079 Homoserine dehydrogenase n=1 Tax=Zea mays RepID... 135 1e-30
UniRef100_P93402 Aspartate kinase-homoserine dehydrogenase n=1 T... 133 7e-30
UniRef100_B9G2N4 Putative uncharacterized protein n=1 Tax=Oryza ... 133 7e-30
UniRef100_Q69LG7 Os09g0294000 protein n=2 Tax=Oryza sativa RepID... 133 7e-30
UniRef100_C5X893 Putative uncharacterized protein Sb02g019450 n=... 131 4e-29
UniRef100_P49080 Homoserine dehydrogenase n=1 Tax=Zea mays RepID... 122 2e-26
UniRef100_A9TAK5 Predicted protein n=1 Tax=Physcomitrella patens... 116 1e-24
UniRef100_A9SKI1 Predicted protein n=1 Tax=Physcomitrella patens... 112 2e-23
UniRef100_B4SGC5 Aspartate kinase n=1 Tax=Pelodictyon phaeoclath... 64 9e-09
UniRef100_A8J1S7 Bifunctional aspartate kinase/homoserine dehydr... 63 1e-08
UniRef100_A4AQY1 Aspartokinase/homoserine dehydrogenase n=1 Tax=... 59 2e-07
UniRef100_Q3B1F6 Homoserine dehydrogenase / aspartate kinase n=1... 59 2e-07
UniRef100_A7Q808 Chromosome chr18 scaffold_61, whole genome shot... 59 2e-07
UniRef100_Q11YZ9 Bifunctional protein: aspartokinase I; homoseri... 58 4e-07
UniRef100_B3QZD1 Aspartate kinase n=1 Tax=Chloroherpeton thalass... 58 4e-07
UniRef100_Q2S4Q2 Bifunctional aspartokinase/homoserine dehydroge... 57 1e-06
UniRef100_B3EHJ1 Aspartate kinase n=1 Tax=Chlorobium limicola DS... 56 2e-06
UniRef100_C5LZJ0 Aspartokinase n=1 Tax=Perkinsus marinus ATCC 50... 56 2e-06
UniRef100_Q8KAX1 Aspartokinase/homoserine dehydrogenase n=1 Tax=... 55 4e-06
UniRef100_C1MP14 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 4e-06
UniRef100_C5LXT8 Aspartokinase (Fragment) n=1 Tax=Perkinsus mari... 54 7e-06
UniRef100_Q9F1V0 Aspartate kinase-homoserine dehydrogenase (Frag... 54 1e-05
UniRef100_A9T456 Aspartokinase n=1 Tax=Physcomitrella patens sub... 54 1e-05
UniRef100_A9NUI1 Aspartokinase n=1 Tax=Picea sitchensis RepID=A9... 54 1e-05