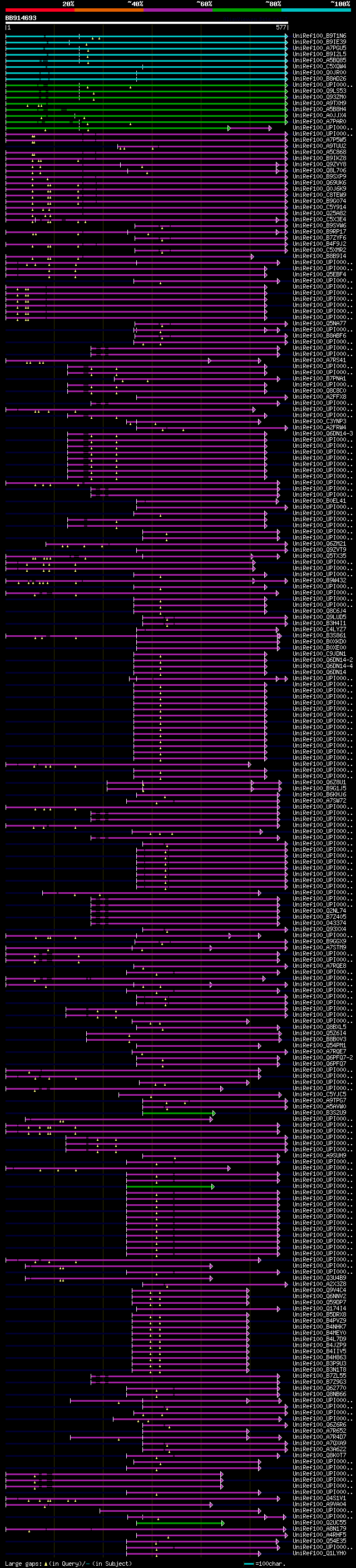
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB914693 RCE14283
(577 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9T1N6 Synaptotagmin, putative n=1 Tax=Ricinus communi... 307 3e-82
UniRef100_B9IE39 Plant synaptotagmin n=1 Tax=Populus trichocarpa... 298 2e-79
UniRef100_A7PGU5 Chromosome chr17 scaffold_16, whole genome shot... 293 8e-78
UniRef100_B9I2L5 Plant synaptotagmin n=1 Tax=Populus trichocarpa... 288 3e-76
UniRef100_A5BQ85 Putative uncharacterized protein n=1 Tax=Vitis ... 283 5e-75
UniRef100_C5XQW4 Plant synaptotagmin n=1 Tax=Sorghum bicolor Rep... 264 4e-69
UniRef100_Q0JR00 Os01g0128800 protein (Fragment) n=1 Tax=Oryza s... 260 5e-68
UniRef100_B8AD26 Putative uncharacterized protein n=1 Tax=Oryza ... 260 5e-68
UniRef100_UPI0001984C04 PREDICTED: hypothetical protein n=1 Tax=... 251 2e-65
UniRef100_Q9LS53 Genomic DNA, chromosome 3, P1 clone: MYF24 n=1 ... 232 2e-59
UniRef100_Q93ZM0 AT3g18370/MYF24_8 n=1 Tax=Arabidopsis thaliana ... 232 2e-59
UniRef100_A9TXH9 Predicted protein n=1 Tax=Physcomitrella patens... 229 2e-58
UniRef100_A5B8H4 Putative uncharacterized protein n=1 Tax=Vitis ... 229 2e-58
UniRef100_A0JJX4 NTMC2Type3.1 protein (Fragment) n=1 Tax=Physcom... 229 2e-58
UniRef100_A7PAR0 Chromosome chr14 scaffold_9, whole genome shotg... 197 5e-49
UniRef100_UPI0001984F2B PREDICTED: hypothetical protein n=1 Tax=... 176 9e-43
UniRef100_UPI000198311D PREDICTED: hypothetical protein n=1 Tax=... 84 1e-14
UniRef100_A7P5W5 Chromosome chr4 scaffold_6, whole genome shotgu... 84 1e-14
UniRef100_A9TUU2 Predicted protein n=1 Tax=Physcomitrella patens... 82 2e-14
UniRef100_A5C868 Putative uncharacterized protein n=1 Tax=Vitis ... 82 2e-14
UniRef100_B9IKZ8 Plant synaptotagmin n=1 Tax=Populus trichocarpa... 76 2e-12
UniRef100_Q9ZVY8 T25N20.15 n=1 Tax=Arabidopsis thaliana RepID=Q9... 75 3e-12
UniRef100_Q8L706 Ca2+-dependent lipid-binding protein, putative ... 75 3e-12
UniRef100_B9SXP9 Calcium lipid binding protein, putative n=1 Tax... 75 3e-12
UniRef100_Q69UK6 Putative C2 domain-containing protein n=1 Tax=O... 74 1e-11
UniRef100_Q0J6K9 Os08g0300200 protein n=2 Tax=Oryza sativa Japon... 74 1e-11
UniRef100_C8TEW9 Putative CLB1 protein n=1 Tax=Oryza sativa Indi... 74 1e-11
UniRef100_B9G074 Putative uncharacterized protein n=1 Tax=Oryza ... 74 1e-11
UniRef100_C5Y914 Putative uncharacterized protein Sb06g030540 n=... 73 2e-11
UniRef100_Q25A82 H0413E07.12 protein n=3 Tax=Oryza sativa RepID=... 72 3e-11
UniRef100_C5X3E4 Plant synaptotagmin n=1 Tax=Sorghum bicolor Rep... 70 9e-11
UniRef100_B9SVW6 Putative uncharacterized protein n=1 Tax=Ricinu... 70 9e-11
UniRef100_B9RP17 Calcium lipid binding protein, putative n=1 Tax... 70 1e-10
UniRef100_B7ZYF6 Putative uncharacterized protein n=1 Tax=Zea ma... 70 2e-10
UniRef100_B4F9J2 Lipid binding protein n=1 Tax=Zea mays RepID=B4... 70 2e-10
UniRef100_C5XMR2 Integral membrane single C2 domain protein n=1 ... 67 8e-10
UniRef100_B8B9I4 Putative uncharacterized protein n=1 Tax=Oryza ... 67 8e-10
UniRef100_UPI000194DFD0 PREDICTED: multiple C2 domains, transmem... 67 1e-09
UniRef100_UPI000069E697 MGC108303 protein. n=1 Tax=Xenopus (Silu... 67 1e-09
UniRef100_Q5EBF4 MGC108303 protein n=1 Tax=Xenopus (Silurana) tr... 67 1e-09
UniRef100_UPI00017B15FA UPI00017B15FA related cluster n=1 Tax=Te... 67 1e-09
UniRef100_UPI00016E2724 UPI00016E2724 related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_UPI00016E2723 UPI00016E2723 related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_UPI00016E2722 UPI00016E2722 related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_UPI00016E2721 UPI00016E2721 related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_UPI00016E2720 UPI00016E2720 related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_UPI00016E271F UPI00016E271F related cluster n=1 Tax=Ta... 67 1e-09
UniRef100_Q5NA77 Os01g0242600 protein n=2 Tax=Oryza sativa Japon... 66 2e-09
UniRef100_UPI0000E8199F PREDICTED: hypothetical protein n=1 Tax=... 66 2e-09
UniRef100_B8ABF6 Putative uncharacterized protein n=1 Tax=Oryza ... 66 2e-09
UniRef100_UPI0000E4A1E6 PREDICTED: similar to synaptotagmin, put... 66 2e-09
UniRef100_UPI00005A0EE3 PREDICTED: similar to RAS p21 protein ac... 66 2e-09
UniRef100_UPI0000EB32C8 UPI0000EB32C8 related cluster n=1 Tax=Ca... 66 2e-09
UniRef100_A7RS41 Predicted protein (Fragment) n=1 Tax=Nematostel... 66 2e-09
UniRef100_UPI0000DA2031 PREDICTED: similar to multiple C2-domain... 65 3e-09
UniRef100_UPI0001B7B9E6 UPI0001B7B9E6 related cluster n=1 Tax=Ra... 65 3e-09
UniRef100_B7PNA1 Multiple C2 and transmembrane domain-containing... 65 3e-09
UniRef100_UPI00001C527D multiple C2 domains, transmembrane 1 n=1... 65 4e-09
UniRef100_Q8C8C0 Putative uncharacterized protein n=1 Tax=Mus mu... 65 4e-09
UniRef100_A2FFX8 Putative uncharacterized protein n=1 Tax=Tricho... 65 5e-09
UniRef100_UPI0001796EAD PREDICTED: RAS p21 protein activator 4 n... 64 6e-09
UniRef100_UPI000175FD85 PREDICTED: similar to multiple C2 domain... 64 6e-09
UniRef100_UPI00015E0714 multiple C2 domains, transmembrane 1 iso... 64 6e-09
UniRef100_C3YNP3 Putative uncharacterized protein n=1 Tax=Branch... 64 6e-09
UniRef100_A2FRW4 C2 domain containing protein n=1 Tax=Trichomona... 64 6e-09
UniRef100_Q6DN14-3 Isoform 3 of Multiple C2 and transmembrane do... 64 6e-09
UniRef100_UPI0000E207CD PREDICTED: hypothetical protein isoform ... 64 8e-09
UniRef100_UPI0000E207CB PREDICTED: multiple C2-domains with two ... 64 8e-09
UniRef100_UPI0000E207CA PREDICTED: multiple C2-domains with two ... 64 8e-09
UniRef100_UPI0000E207C9 PREDICTED: hypothetical protein isoform ... 64 8e-09
UniRef100_UPI0000D9B551 PREDICTED: similar to multiple C2-domain... 64 8e-09
UniRef100_UPI0000D9B550 PREDICTED: similar to multiple C2-domain... 64 8e-09
UniRef100_UPI0000D9B54F PREDICTED: similar to multiple C2-domain... 64 8e-09
UniRef100_UPI0000D562CD PREDICTED: similar to AGAP007646-PA n=1 ... 64 8e-09
UniRef100_UPI00005C0096 PREDICTED: similar to Ras GTPase-activat... 64 8e-09
UniRef100_UPI000179CDCA UPI000179CDCA related cluster n=1 Tax=Bo... 64 8e-09
UniRef100_B0EL41 Circumsporozoite protein, putative n=1 Tax=Enta... 64 8e-09
UniRef100_UPI0001738F89 protein binding / zinc ion binding n=1 T... 64 1e-08
UniRef100_UPI000155455C PREDICTED: similar to MCTP1L n=1 Tax=Orn... 64 1e-08
UniRef100_UPI00005A05CA PREDICTED: similar to multiple C2-domain... 64 1e-08
UniRef100_UPI00005A05C7 PREDICTED: similar to multiple C2-domain... 64 1e-08
UniRef100_UPI0001B7998F UPI0001B7998F related cluster n=1 Tax=Ra... 64 1e-08
UniRef100_UPI000154EECA similar to Rasa4 protein (predicted) (RG... 64 1e-08
UniRef100_Q6ZM21 Novel protein similar to mouse and human membra... 64 1e-08
UniRef100_Q9ZVT9 F15K9.2 protein n=1 Tax=Arabidopsis thaliana Re... 64 1e-08
UniRef100_Q5TX35 AGAP007646-PA n=1 Tax=Anopheles gambiae RepID=Q... 64 1e-08
UniRef100_UPI0001A2BFF9 UPI0001A2BFF9 related cluster n=1 Tax=Da... 63 2e-08
UniRef100_UPI0001A2BF75 UPI0001A2BF75 related cluster n=1 Tax=Da... 63 2e-08
UniRef100_UPI0001B7B9E7 UPI0001B7B9E7 related cluster n=1 Tax=Ra... 63 2e-08
UniRef100_B9W432 Putative C2 domain containing protein (Fragment... 63 2e-08
UniRef100_UPI0000F2C539 PREDICTED: similar to MCTP1L n=1 Tax=Mon... 62 2e-08
UniRef100_UPI0001A2D104 RasGAP-activating-like protein 1. n=1 Ta... 62 2e-08
UniRef100_UPI000021505E multiple C2 domains, transmembrane 1 n=1... 62 2e-08
UniRef100_UPI0000D6165C multiple C2 domains, transmembrane 1 iso... 62 2e-08
UniRef100_Q8C6J4 Putative uncharacterized protein (Fragment) n=1... 62 2e-08
UniRef100_Q9LUD5 Genomic DNA, chromosome 3, P1 clone: MIE1 n=1 T... 62 2e-08
UniRef100_B3H4I1 Uncharacterized protein At3g14590.2 n=2 Tax=Ara... 62 2e-08
UniRef100_C4LYZ7 C2 domain containing protein n=1 Tax=Entamoeba ... 62 2e-08
UniRef100_B3S861 Putative uncharacterized protein n=1 Tax=Tricho... 62 2e-08
UniRef100_B0XKD0 Putative uncharacterized protein n=1 Tax=Culex ... 62 2e-08
UniRef100_B0XE00 Putative uncharacterized protein n=1 Tax=Culex ... 62 2e-08
UniRef100_C9JDN1 Putative uncharacterized protein MCTP1 (Fragmen... 62 2e-08
UniRef100_Q6DN14-2 Isoform 2 of Multiple C2 and transmembrane do... 62 2e-08
UniRef100_Q6DN14-4 Isoform 4 of Multiple C2 and transmembrane do... 62 2e-08
UniRef100_Q6DN14 Multiple C2 and transmembrane domain-containing... 62 2e-08
UniRef100_UPI0001925075 PREDICTED: similar to predicted protein,... 62 3e-08
UniRef100_UPI0000E207CC PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000E207C8 PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000E207C7 PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000E207C6 PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000E207C5 PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000E207C4 PREDICTED: multiple C2-domains with two ... 62 3e-08
UniRef100_UPI0000D9B553 PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI0000D9B54E PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI0000D9B54D PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI00005A05CC PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI00005A05CB PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI00005A05C9 PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI00005A05C8 PREDICTED: similar to multiple C2-domain... 62 3e-08
UniRef100_UPI00017B27E5 UPI00017B27E5 related cluster n=1 Tax=Te... 62 3e-08
UniRef100_UPI0000EB414D multiple C2 domains, transmembrane 1 iso... 62 3e-08
UniRef100_UPI0000EB414C multiple C2 domains, transmembrane 1 iso... 62 3e-08
UniRef100_Q6Z8U1 C2 domain/GRAM domain-containing protein-like n... 62 3e-08
UniRef100_B9G1J5 Putative uncharacterized protein n=2 Tax=Oryza ... 62 3e-08
UniRef100_B6KHJ6 C2 domain-containing protein n=3 Tax=Toxoplasma... 62 3e-08
UniRef100_A7SW72 Predicted protein n=1 Tax=Nematostella vectensi... 62 3e-08
UniRef100_UPI0000DB725F PREDICTED: similar to multiple C2 domain... 62 4e-08
UniRef100_UPI0001A5C53A PREDICTED: similar to calcium-promoted R... 61 5e-08
UniRef100_UPI0000ED4E86 RAS p21 protein activator 4 isoform 2 n=... 61 5e-08
UniRef100_UPI0000E4A99C PREDICTED: hypothetical protein n=1 Tax=... 61 5e-08
UniRef100_UPI000051A58C PREDICTED: similar to Syt7 CG2381-PG, is... 61 5e-08
UniRef100_UPI000020F7DE RAS p21 protein activator 4 isoform 1 n=... 61 5e-08
UniRef100_UPI0000163123 NTMC2T6.1 n=1 Tax=Arabidopsis thaliana R... 61 5e-08
UniRef100_UPI00006A22D4 UPI00006A22D4 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI000069EB17 UPI000069EB17 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI000069EB16 UPI000069EB16 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI000069EB15 UPI000069EB15 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI00004D1DCA UPI00004D1DCA related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI00004D1DC9 UPI00004D1DC9 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI00004D1DC8 UPI00004D1DC8 related cluster n=1 Tax=Xe... 61 5e-08
UniRef100_UPI00017B27E7 UPI00017B27E7 related cluster n=1 Tax=Te... 61 5e-08
UniRef100_UPI0001AE70B9 UPI0001AE70B9 related cluster n=1 Tax=Ho... 61 5e-08
UniRef100_UPI0001A5C53B PREDICTED: similar to calcium-promoted R... 61 5e-08
UniRef100_Q2NL74 RAS p21 protein activator 4 n=1 Tax=Homo sapien... 61 5e-08
UniRef100_B7Z405 cDNA FLJ50019, highly similar to Ras GTPase-act... 61 5e-08
UniRef100_O43374 Ras GTPase-activating protein 4 n=2 Tax=Homo sa... 61 5e-08
UniRef100_Q93XX4 C2 domain-containing protein At1g53590 n=1 Tax=... 61 5e-08
UniRef100_UPI000185F37E hypothetical protein BRAFLDRAFT_65294 n=... 61 7e-08
UniRef100_B9GGX9 Integral membrane single C2 domain protein n=1 ... 61 7e-08
UniRef100_A7STM9 Predicted protein n=1 Tax=Nematostella vectensi... 61 7e-08
UniRef100_UPI0000E81348 PREDICTED: similar to Ras GTPase-activat... 60 9e-08
UniRef100_UPI0000ECA37D UPI0000ECA37D related cluster n=1 Tax=Ga... 60 9e-08
UniRef100_A7RQE8 Predicted protein n=1 Tax=Nematostella vectensi... 60 9e-08
UniRef100_UPI000175F2FD PREDICTED: similar to unc-13 homolog B n... 60 1e-07
UniRef100_UPI0000F2C9C5 PREDICTED: similar to RasGAP-activating-... 60 1e-07
UniRef100_UPI0000D5624F PREDICTED: similar to synaptotagmin, put... 60 1e-07
UniRef100_UPI0001A2BA8B UPI0001A2BA8B related cluster n=1 Tax=Da... 60 1e-07
UniRef100_UPI00004D1DCB UPI00004D1DCB related cluster n=1 Tax=Xe... 60 1e-07
UniRef100_UPI00004D1DC6 UPI00004D1DC6 related cluster n=1 Tax=Xe... 60 1e-07
UniRef100_UPI00017B271A UPI00017B271A related cluster n=1 Tax=Te... 60 1e-07
UniRef100_UPI00017B2719 UPI00017B2719 related cluster n=1 Tax=Te... 60 1e-07
UniRef100_UPI0000D57736 PREDICTED: similar to Syt7 CG2381-PB, pa... 60 2e-07
UniRef100_Q8BXL5 Putative uncharacterized protein n=1 Tax=Mus mu... 60 2e-07
UniRef100_Q5Z6I4 Os06g0297800 protein n=1 Tax=Oryza sativa Japon... 60 2e-07
UniRef100_B8B0V3 Putative uncharacterized protein n=1 Tax=Oryza ... 60 2e-07
UniRef100_Q54PM1 C2 domain-containing protein n=1 Tax=Dictyostel... 60 2e-07
UniRef100_A7RQE7 Predicted protein n=1 Tax=Nematostella vectensi... 60 2e-07
UniRef100_Q6PFQ7-2 Isoform 2 of Ras GTPase-activating protein 4 ... 60 2e-07
UniRef100_Q6PFQ7 Ras GTPase-activating protein 4 n=1 Tax=Mus mus... 60 2e-07
UniRef100_UPI000187E072 hypothetical protein MPER_04373 n=1 Tax=... 59 2e-07
UniRef100_UPI000175F7D3 PREDICTED: similar to multiple C2 domain... 59 2e-07
UniRef100_UPI00015B4343 PREDICTED: similar to CG2381-PA n=1 Tax=... 59 2e-07
UniRef100_UPI00016E8C26 UPI00016E8C26 related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_C5YJC5 Putative uncharacterized protein Sb07g028720 n=... 59 2e-07
UniRef100_A9TPG7 Predicted protein n=1 Tax=Physcomitrella patens... 59 2e-07
UniRef100_A5AVW0 Putative uncharacterized protein n=1 Tax=Vitis ... 59 2e-07
UniRef100_B3S2U9 Putative uncharacterized protein n=1 Tax=Tricho... 59 2e-07
UniRef100_UPI0000F4ABE9 multiple C2 domains, transmembrane 1 n=1... 59 3e-07
UniRef100_UPI00016E83D7 UPI00016E83D7 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E83D6 UPI00016E83D6 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E1F53 UPI00016E1F53 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E1F52 UPI00016E1F52 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E1F50 UPI00016E1F50 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_A9SUH9 Predicted protein n=1 Tax=Physcomitrella patens... 59 3e-07
UniRef100_UPI00017C2EF7 PREDICTED: similar to unc-13 homolog C n... 59 4e-07
UniRef100_UPI0001561060 PREDICTED: similar to family with sequen... 59 4e-07
UniRef100_UPI000155D8EE PREDICTED: unc-13 homolog C (C. elegans)... 59 4e-07
UniRef100_UPI0000F2B180 PREDICTED: similar to unc-13 homolog C n... 59 4e-07
UniRef100_UPI0000E46814 PREDICTED: similar to UNC13 (C. elegans)... 59 4e-07
UniRef100_UPI0000E23CCA PREDICTED: similar to Munc13-3 n=1 Tax=P... 59 4e-07
UniRef100_UPI0000D9B936 PREDICTED: similar to unc-13 homolog C i... 59 4e-07
UniRef100_UPI0000D9B935 PREDICTED: similar to unc-13 homolog C i... 59 4e-07
UniRef100_UPI0000D9B934 PREDICTED: similar to unc-13 homolog C i... 59 4e-07
UniRef100_UPI0000D9B933 PREDICTED: similar to unc-13 homolog C i... 59 4e-07
UniRef100_UPI00005A51D3 PREDICTED: similar to Unc-13 homolog C (... 59 4e-07
UniRef100_UPI00005A51D2 PREDICTED: similar to unc-13 homolog C i... 59 4e-07
UniRef100_UPI00005A51D1 PREDICTED: similar to Unc-13 homolog C (... 59 4e-07
UniRef100_UPI00005A51D0 PREDICTED: similar to Unc-13 homolog C (... 59 4e-07
UniRef100_UPI00005A51CF PREDICTED: similar to Unc-13 homolog C (... 59 4e-07
UniRef100_UPI00005A51CE PREDICTED: similar to Unc-13 homolog C (... 59 4e-07
UniRef100_UPI0001A2DC19 UPI0001A2DC19 related cluster n=1 Tax=Da... 59 4e-07
UniRef100_UPI0001B7B9E8 UPI0001B7B9E8 related cluster n=1 Tax=Ra... 59 4e-07
UniRef100_UPI000179E5FA Unc-13 homolog C (Munc13-3). n=1 Tax=Bos... 59 4e-07
UniRef100_Q3U4B9 Putative uncharacterized protein n=1 Tax=Mus mu... 59 4e-07
UniRef100_A2X3Z8 Putative uncharacterized protein n=1 Tax=Oryza ... 59 4e-07
UniRef100_Q9V4C4 Syt7, isoform A n=1 Tax=Drosophila melanogaster... 59 4e-07
UniRef100_Q6NNV2 Syt7, isoform B n=1 Tax=Drosophila melanogaster... 59 4e-07
UniRef100_Q59DP7 Syt7, isoform F n=1 Tax=Drosophila melanogaster... 59 4e-07
UniRef100_Q174I4 Putative uncharacterized protein (Fragment) n=1... 59 4e-07
UniRef100_B5DRX8 GA22513 (Fragment) n=1 Tax=Drosophila pseudoobs... 59 4e-07
UniRef100_B4PVZ9 GE14519 n=1 Tax=Drosophila yakuba RepID=B4PVZ9_... 59 4e-07
UniRef100_B4NHK7 GK13680 n=1 Tax=Drosophila willistoni RepID=B4N... 59 4e-07
UniRef100_B4MEY0 GJ12969 n=1 Tax=Drosophila virilis RepID=B4MEY0... 59 4e-07
UniRef100_B4L7D9 GI14040 n=1 Tax=Drosophila mojavensis RepID=B4L... 59 4e-07
UniRef100_B4JZP9 GH23960 n=1 Tax=Drosophila grimshawi RepID=B4JZ... 59 4e-07
UniRef100_B4IIV5 GM26786 n=1 Tax=Drosophila sechellia RepID=B4II... 59 4e-07
UniRef100_B4H863 GL18166 n=1 Tax=Drosophila persimilis RepID=B4H... 59 4e-07
UniRef100_B3P9U3 GG16453 n=1 Tax=Drosophila erecta RepID=B3P9U3_... 59 4e-07
UniRef100_B3N1T8 GF20608 n=1 Tax=Drosophila ananassae RepID=B3N1... 59 4e-07
UniRef100_B7ZL55 RAS p21 protein activator 4 n=1 Tax=Homo sapien... 59 4e-07
UniRef100_B7Z9G3 cDNA, FLJ78827, highly similar to Ras GTPase-ac... 59 4e-07
UniRef100_Q62770 Protein unc-13 homolog C n=1 Tax=Rattus norvegi... 59 4e-07
UniRef100_Q8NB66 Protein unc-13 homolog C n=1 Tax=Homo sapiens R... 59 4e-07
UniRef100_UPI0001985F6E PREDICTED: hypothetical protein n=1 Tax=... 58 5e-07
UniRef100_UPI0001985801 PREDICTED: hypothetical protein n=1 Tax=... 58 5e-07
UniRef100_UPI000186ED37 conserved hypothetical protein n=1 Tax=P... 58 5e-07
UniRef100_UPI0001797D6E PREDICTED: similar to Synaptotagmin-2 (S... 58 5e-07
UniRef100_Q6Z6R6 Os02g0313700 protein n=1 Tax=Oryza sativa Japon... 58 5e-07
UniRef100_A7R652 Chromosome undetermined scaffold_1169, whole ge... 58 5e-07
UniRef100_A7R4D7 Chromosome undetermined scaffold_658, whole gen... 58 5e-07
UniRef100_A7QXA9 Chromosome chr19 scaffold_218, whole genome sho... 58 5e-07
UniRef100_A3A622 Putative uncharacterized protein n=1 Tax=Oryza ... 58 5e-07
UniRef100_Q8K0T7 Protein unc-13 homolog C n=1 Tax=Mus musculus R... 58 5e-07
UniRef100_UPI00017B27E6 UPI00017B27E6 related cluster n=1 Tax=Te... 58 6e-07
UniRef100_UPI0001B7AA31 Unc-13 homolog C (Munc13-3). n=1 Tax=Rat... 58 6e-07
UniRef100_UPI00016E8C07 UPI00016E8C07 related cluster n=1 Tax=Ta... 58 6e-07
UniRef100_UPI00016E8C06 UPI00016E8C06 related cluster n=1 Tax=Ta... 58 6e-07
UniRef100_UPI00016E8C05 UPI00016E8C05 related cluster n=1 Tax=Ta... 58 6e-07
UniRef100_UPI0000EB06A2 UPI0000EB06A2 related cluster n=1 Tax=Ca... 58 6e-07
UniRef100_Q4S1V1 Chromosome undetermined SCAF14764, whole genome... 58 6e-07
UniRef100_A9VA04 Predicted protein n=1 Tax=Monosiga brevicollis ... 58 6e-07
UniRef100_UPI0001925A7B PREDICTED: similar to predicted protein ... 57 8e-07
UniRef100_UPI000069E5AC multiple C2 domains, transmembrane 2 n=1... 57 8e-07
UniRef100_Q2UC55 Phosphatidylserine decarboxylase n=1 Tax=Asperg... 57 8e-07
UniRef100_A8N179 Putative uncharacterized protein n=1 Tax=Coprin... 57 8e-07
UniRef100_A4RHF5 Putative uncharacterized protein n=1 Tax=Magnap... 57 8e-07
UniRef100_Q54E35 Rho GTPase-activating protein gacEE n=1 Tax=Dic... 57 8e-07
UniRef100_UPI0000E80C85 PREDICTED: similar to Munc13-3 n=1 Tax=G... 57 1e-06
UniRef100_Q1LYH0 Novel protein similar to vertebrate unc-13 homo... 57 1e-06
UniRef100_UPI00017B4113 UPI00017B4113 related cluster n=1 Tax=Te... 57 1e-06
UniRef100_UPI00017B4112 UPI00017B4112 related cluster n=1 Tax=Te... 57 1e-06
UniRef100_UPI00017B1278 UPI00017B1278 related cluster n=1 Tax=Te... 57 1e-06
UniRef100_UPI00016E8660 UPI00016E8660 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E865F UPI00016E865F related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E8639 UPI00016E8639 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E8638 UPI00016E8638 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E2FD4 UPI00016E2FD4 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E2FD3 UPI00016E2FD3 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E2FD2 UPI00016E2FD2 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E2DD1 UPI00016E2DD1 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI0000ECAF25 Unc-13 homolog C (Munc13-3). n=1 Tax=Gal... 57 1e-06
UniRef100_Q1LXS6 Novel protein similar to vertebrate unc-13 homo... 57 1e-06
UniRef100_C5Z915 Putative uncharacterized protein Sb10g010850 n=... 57 1e-06
UniRef100_A9SRR8 Predicted protein n=1 Tax=Physcomitrella patens... 57 1e-06
UniRef100_O15755 C2 domain-containing protein n=1 Tax=Dictyostel... 57 1e-06
UniRef100_B3RXZ7 Predicted protein n=1 Tax=Trichoplax adhaerens ... 57 1e-06
UniRef100_A2FNQ6 XYPPX repeat family protein n=1 Tax=Trichomonas... 57 1e-06
UniRef100_A2E8X1 C2 domain containing protein (Fragment) n=1 Tax... 57 1e-06
UniRef100_B8N754 Phosphatidylserine decarboxylase Psd2, putative... 57 1e-06
UniRef100_A6SQ97 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_Q5FWL4 Extended synaptotagmin-2-A n=1 Tax=Xenopus laev... 57 1e-06
UniRef100_UPI000194DC00 PREDICTED: similar to unc-13 homolog B (... 57 1e-06
UniRef100_UPI00015565A7 PREDICTED: similar to unc-13 homolog C, ... 57 1e-06
UniRef100_UPI0000E218D2 PREDICTED: similar to RAS p21 protein ac... 57 1e-06
UniRef100_UPI0000E21682 PREDICTED: hypothetical protein n=1 Tax=... 57 1e-06
UniRef100_UPI0000D9CD0C PREDICTED: similar to family with sequen... 57 1e-06
UniRef100_UPI0000D9CD0B PREDICTED: similar to family with sequen... 57 1e-06
UniRef100_UPI0000D9CD0A PREDICTED: similar to family with sequen... 57 1e-06
UniRef100_UPI0000D575D9 PREDICTED: similar to unc-13 CG2999-PC n... 57 1e-06
UniRef100_Q3Y418 Synaptotagmin protein 2 (Fragment) n=2 Tax=Caen... 57 1e-06
UniRef100_UPI00016E83DA UPI00016E83DA related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E83D9 UPI00016E83D9 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E83D8 UPI00016E83D8 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_UPI00016E83D5 UPI00016E83D5 related cluster n=1 Tax=Ta... 57 1e-06
UniRef100_Q2A9R2 C2 domain containing protein n=1 Tax=Brassica o... 57 1e-06
UniRef100_A9TNU8 Predicted protein n=1 Tax=Physcomitrella patens... 57 1e-06
UniRef100_B4JG26 GH18195 n=1 Tax=Drosophila grimshawi RepID=B4JG... 57 1e-06
UniRef100_A8PJY3 C2 domain containing protein n=1 Tax=Brugia mal... 57 1e-06
UniRef100_Q12466 Tricalbin-1 n=1 Tax=Saccharomyces cerevisiae Re... 57 1e-06
UniRef100_Q5RAG2 Extended synaptotagmin-1 n=1 Tax=Pongo abelii R... 57 1e-06
UniRef100_UPI0000E47FEC PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000DB75C1 PREDICTED: similar to CG6454-PA, isoform... 56 2e-06
UniRef100_UPI000184A469 Unc-13 homolog C (Munc13-3). n=1 Tax=Xen... 56 2e-06
UniRef100_UPI000069F9C6 Unc-13 homolog C (Munc13-3). n=1 Tax=Xen... 56 2e-06
UniRef100_UPI00017B1173 UPI00017B1173 related cluster n=1 Tax=Te... 56 2e-06
UniRef100_UPI00017B1150 UPI00017B1150 related cluster n=1 Tax=Te... 56 2e-06
UniRef100_UPI00016E2BBB UPI00016E2BBB related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_UPI00016E2B9A UPI00016E2B9A related cluster n=1 Tax=Ta... 56 2e-06
UniRef100_Q4RV37 Chromosome 15 SCAF14992, whole genome shotgun s... 56 2e-06
UniRef100_Q9FIK8 Similarity to GTPase activating protein n=1 Tax... 56 2e-06
UniRef100_C5Z714 Integral membrane single C2 domain protein n=1 ... 56 2e-06
UniRef100_A9RX14 Predicted protein n=1 Tax=Physcomitrella patens... 56 2e-06
UniRef100_B4L7A5 GI14058 n=1 Tax=Drosophila mojavensis RepID=B4L... 56 2e-06
UniRef100_B0EHK2 Putative uncharacterized protein n=1 Tax=Entamo... 56 2e-06
UniRef100_B7Z4G1 cDNA FLJ54825, highly similar to Mus musculus m... 56 2e-06
UniRef100_C8ZI48 Tcb1p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 56 2e-06
UniRef100_C5FV51 C2 domain-containing protein n=1 Tax=Microsporu... 56 2e-06
UniRef100_B5VRY2 YOR086Cp-like protein n=1 Tax=Saccharomyces cer... 56 2e-06
UniRef100_A6ZNT7 Conserved protein n=2 Tax=Saccharomyces cerevis... 56 2e-06
UniRef100_UPI000194CFC6 PREDICTED: unc-13 homolog C (C. elegans)... 56 2e-06
UniRef100_UPI000186E79F unc-13, putative n=1 Tax=Pediculus human... 56 2e-06
UniRef100_UPI00017C315C PREDICTED: similar to Protein unc-13 hom... 56 2e-06
UniRef100_UPI000155C669 PREDICTED: similar to munc 13-1 n=1 Tax=... 56 2e-06
UniRef100_UPI00005A3C13 PREDICTED: similar to Unc-13 homolog A (... 56 2e-06
UniRef100_UPI0000428F99 unc-13 homolog A n=1 Tax=Mus musculus Re... 56 2e-06
UniRef100_UPI000184A43B Unc-13 homolog A (Munc13-1). n=1 Tax=Xen... 56 2e-06
UniRef100_UPI00006A093D Unc-13 homolog A (Munc13-1). n=1 Tax=Xen... 56 2e-06
UniRef100_UPI00015DEEDF unc-13 homolog A (C. elegans) n=1 Tax=Mu... 56 2e-06
UniRef100_UPI0000D8AD8C unc-13 homolog A (C. elegans) n=1 Tax=Mu... 56 2e-06
UniRef100_UPI0000DD84DC unc-13 homolog A n=1 Tax=Homo sapiens Re... 56 2e-06
UniRef100_UPI0000EB1A94 Unc-13 homolog A (Munc13-1). n=1 Tax=Can... 56 2e-06
UniRef100_UPI000179E7E7 UPI000179E7E7 related cluster n=1 Tax=Bo... 56 2e-06
UniRef100_Q4SDC9 Chromosome 1 SCAF14640, whole genome shotgun se... 56 2e-06
UniRef100_C4M0W7 C2 domain protein, putative n=1 Tax=Entamoeba h... 56 2e-06
UniRef100_C4LYI3 Putative uncharacterized protein n=1 Tax=Entamo... 56 2e-06
UniRef100_B4KB48 GI10148 n=1 Tax=Drosophila mojavensis RepID=B4K... 56 2e-06
UniRef100_A2DBY3 C2 domain containing protein n=1 Tax=Trichomona... 56 2e-06
UniRef100_C9JCN4 Putative uncharacterized protein UNC13A (Fragme... 56 2e-06
UniRef100_Q4WYR4 Phosphatidylserine decarboxylase Psd2, putative... 56 2e-06
UniRef100_B0Y097 Phosphatidylserine decarboxylase Psd2, putative... 56 2e-06
UniRef100_Q62768-2 Isoform 2 of Protein unc-13 homolog A n=1 Tax... 56 2e-06
UniRef100_Q62768-3 Isoform 3 of Protein unc-13 homolog A n=1 Tax... 56 2e-06
UniRef100_Q62768 Protein unc-13 homolog A n=1 Tax=Rattus norvegi... 56 2e-06
UniRef100_Q4KUS2 Protein unc-13 homolog A n=1 Tax=Mus musculus R... 56 2e-06
UniRef100_Q9UPW8 Protein unc-13 homolog A n=1 Tax=Homo sapiens R... 56 2e-06
UniRef100_UPI0001864A41 hypothetical protein BRAFLDRAFT_83913 n=... 55 3e-06
UniRef100_UPI0000F2C4F8 PREDICTED: similar to RAS p21 protein ac... 55 3e-06
UniRef100_UPI000069F733 UPI000069F733 related cluster n=1 Tax=Xe... 55 3e-06
UniRef100_UPI000069DDB3 Synaptotagmin-2 (Synaptotagmin II) (SytI... 55 3e-06
UniRef100_UPI00016E3660 UPI00016E3660 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI00016E365F UPI00016E365F related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI00016E365E UPI00016E365E related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI00016E364A UPI00016E364A related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_Q6DC56 Si:ch211-219a4.7 protein (Fragment) n=1 Tax=Dan... 55 3e-06
UniRef100_B0UYL4 Novel protein similar to vertebrate family with... 55 3e-06
UniRef100_Q6Z7A3 Os02g0199800 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_Q69MT7 Os09g0516900 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_B9G4K6 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_B9F3V9 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_B8LQM3 Putative uncharacterized protein n=1 Tax=Picea ... 55 3e-06
UniRef100_B8BDJ5 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_B8ADT2 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_A6N0H3 Calcium lipid binding protein like (Fragment) n... 55 3e-06
UniRef100_C3ZF76 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_B3RRI8 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_A8XLH0 C. briggsae CBR-SNT-2 protein n=2 Tax=Caenorhab... 55 3e-06
UniRef100_A7SX17 Predicted protein (Fragment) n=1 Tax=Nematostel... 55 3e-06
UniRef100_Q5KKG1 Transmembrane protein, putative n=1 Tax=Filobas... 55 3e-06
UniRef100_Q55VS8 Putative uncharacterized protein n=1 Tax=Filoba... 55 3e-06
UniRef100_B0CP36 Predicted protein n=1 Tax=Laccaria bicolor S238... 55 3e-06
UniRef100_UPI0000E230FD PREDICTED: family with sequence similari... 55 4e-06
UniRef100_UPI0001A2DEBA UPI0001A2DEBA related cluster n=1 Tax=Da... 55 4e-06
UniRef100_UPI00006A1672 fer-1-like 6 n=1 Tax=Xenopus (Silurana) ... 55 4e-06
UniRef100_UPI00006A1671 fer-1-like 6 n=1 Tax=Xenopus (Silurana) ... 55 4e-06
UniRef100_UPI00017B4C9B UPI00017B4C9B related cluster n=1 Tax=Te... 55 4e-06
UniRef100_UPI00017B2718 UPI00017B2718 related cluster n=1 Tax=Te... 55 4e-06
UniRef100_UPI00016E83DB UPI00016E83DB related cluster n=1 Tax=Ta... 55 4e-06
UniRef100_UPI000179EBCB PREDICTED: Bos taurus similar to MCTP1L ... 55 4e-06
UniRef100_Q4T1V0 Chromosome undetermined SCAF10467, whole genome... 55 4e-06
UniRef100_C5X5F0 Putative uncharacterized protein Sb02g029920 n=... 55 4e-06
UniRef100_B9IEY4 Predicted protein n=1 Tax=Populus trichocarpa R... 55 4e-06
UniRef100_Q16WM9 Synaptotagmin, putative (Fragment) n=1 Tax=Aede... 55 4e-06
UniRef100_C6KRL5 Protein ZK524.2f, partially confirmed by transc... 55 4e-06
UniRef100_C3YZN2 Putative uncharacterized protein n=1 Tax=Branch... 55 4e-06
UniRef100_B7QMW4 Munc13-3, putative (Fragment) n=1 Tax=Ixodes sc... 55 4e-06
UniRef100_B0EU86 Synaptotagmin, putative n=1 Tax=Entamoeba dispa... 55 4e-06
UniRef100_A8XEM8 C. briggsae CBR-UNC-13 protein n=1 Tax=Caenorha... 55 4e-06
UniRef100_A2FZS0 C2 domain containing protein n=1 Tax=Trichomona... 55 4e-06
UniRef100_Q0CQJ9 Putative uncharacterized protein n=1 Tax=Asperg... 55 4e-06
UniRef100_B8M4W9 Phosphatidylserine decarboxylase Psd2, putative... 55 4e-06
UniRef100_B8M4W8 Phosphatidylserine decarboxylase Psd2, putative... 55 4e-06
UniRef100_B8M4W7 Phosphatidylserine decarboxylase Psd2, putative... 55 4e-06
UniRef100_B6Q314 Phosphatidylserine decarboxylase Psd2, putative... 55 4e-06
UniRef100_A7EYQ9 Putative uncharacterized protein n=1 Tax=Sclero... 55 4e-06
UniRef100_A2QU82 Catalytic activity: Phosphatidyl-L-serine = Pho... 55 4e-06
UniRef100_A1CL98 Phosphatidylserine decarboxylase n=1 Tax=Asperg... 55 4e-06
UniRef100_Q55A55 Probable serine/threonine-protein kinase DDB_G0... 55 4e-06
UniRef100_P27715-2 Isoform 2 of Phorbol ester/diacylglycerol-bin... 55 4e-06
UniRef100_P27715-7 Isoform 7 of Phorbol ester/diacylglycerol-bin... 55 4e-06
UniRef100_P27715-1 Isoform 1 of Phorbol ester/diacylglycerol-bin... 55 4e-06
UniRef100_P27715 Phorbol ester/diacylglycerol-binding protein un... 55 4e-06
UniRef100_UPI000186E287 conserved hypothetical protein n=1 Tax=P... 55 5e-06
UniRef100_UPI0001797936 PREDICTED: similar to UNC13B protein iso... 55 5e-06
UniRef100_UPI000175F9F2 PREDICTED: family with sequence similari... 55 5e-06
UniRef100_UPI000155C710 PREDICTED: similar to synaptotagmin II -... 55 5e-06
UniRef100_UPI0000F2BB8A PREDICTED: similar to Synaptotagmin-2 (S... 55 5e-06
UniRef100_UPI0000F21142 PREDICTED: similar to Extended-synaptota... 55 5e-06
UniRef100_UPI0000D9DE95 PREDICTED: similar to UNC13 (C. elegans)... 55 5e-06
UniRef100_UPI0000D99E04 PREDICTED: similar to synaptotagmin II n... 55 5e-06
UniRef100_UPI0000D56250 PREDICTED: similar to synaptotagmin, put... 55 5e-06
UniRef100_UPI00005BD2AA PREDICTED: unc-13 homolog B (C. elegans)... 55 5e-06
UniRef100_UPI00005A12ED PREDICTED: similar to Synaptotagmin-2 (S... 55 5e-06
UniRef100_UPI0001A2C1F8 Protein FAM62A (Membrane-bound C2 domain... 55 5e-06
UniRef100_UPI0001B7B4F2 UPI0001B7B4F2 related cluster n=1 Tax=Ra... 55 5e-06
UniRef100_UPI0001B7B4F1 Unc-13 homolog B (Munc13-2). n=1 Tax=Rat... 55 5e-06
UniRef100_UPI0001B7B4F0 UPI0001B7B4F0 related cluster n=1 Tax=Ra... 55 5e-06
UniRef100_UPI0001B7B4EF Unc-13 homolog B (Munc13-2). n=1 Tax=Rat... 55 5e-06
UniRef100_UPI0001B7B4EE UPI0001B7B4EE related cluster n=1 Tax=Ra... 55 5e-06
UniRef100_UPI000050356A Unc-13 homolog B (Munc13-2). n=1 Tax=Rat... 55 5e-06
UniRef100_UPI000019B866 Synaptotagmin-2 (Synaptotagmin II) (SytI... 55 5e-06
UniRef100_UPI0000D65F69 unc-13 homolog B n=1 Tax=Mus musculus Re... 55 5e-06
UniRef100_UPI00016E253C UPI00016E253C related cluster n=1 Tax=Ta... 55 5e-06
UniRef100_UPI00016E2518 UPI00016E2518 related cluster n=1 Tax=Ta... 55 5e-06
UniRef100_UPI00016E2517 UPI00016E2517 related cluster n=1 Tax=Ta... 55 5e-06
UniRef100_UPI00016E2516 UPI00016E2516 related cluster n=1 Tax=Ta... 55 5e-06
UniRef100_UPI00016E2515 UPI00016E2515 related cluster n=1 Tax=Ta... 55 5e-06
UniRef100_UPI0001951270 Synaptotagmin-2 (Synaptotagmin II) (SytI... 55 5e-06
UniRef100_UPI0000EB30F9 Unc-13 homolog B (Munc13-2) (munc13). n=... 55 5e-06
UniRef100_UPI000179EB4F Unc-13 B-like protein n=1 Tax=Bos taurus... 55 5e-06
UniRef100_Q9JM89 Synaptotagmin II (Fragment) n=1 Tax=Mus musculu... 55 5e-06
UniRef100_Q6BCX2 Munc13-1 n=1 Tax=Mus musculus RepID=Q6BCX2_MOUSE 55 5e-06
UniRef100_Q5DTI3 MKIAA4194 protein (Fragment) n=1 Tax=Mus muscul... 55 5e-06
UniRef100_B2RXY6 Unc-13 homolog B (C. elegans) n=1 Tax=Mus muscu... 55 5e-06
UniRef100_B2RXT0 Unc13b protein n=1 Tax=Mus musculus RepID=B2RXT... 55 5e-06
UniRef100_A2AIL5 Unc-13 homolog B (C. elegans) n=1 Tax=Mus muscu... 55 5e-06
UniRef100_A2AG44 Unc-13 homolog B (C. elegans) n=1 Tax=Mus muscu... 55 5e-06
UniRef100_A2AG43 Unc-13 homolog B (C. elegans) n=1 Tax=Mus muscu... 55 5e-06
UniRef100_C1E9K4 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 5e-06
UniRef100_Q9U4K9 UNC-13 n=1 Tax=Drosophila melanogaster RepID=Q9... 55 5e-06
UniRef100_B3KY56 cDNA FLJ46898 fis, clone UTERU3022168, highly s... 55 5e-06
UniRef100_B3KMV5 cDNA FLJ12728 fis, clone NT2RP2000040, highly s... 55 5e-06
UniRef100_A4FU00 SYT2 protein (Fragment) n=1 Tax=Homo sapiens Re... 55 5e-06
UniRef100_Q5B8E2 Putative uncharacterized protein n=1 Tax=Emeric... 55 5e-06
UniRef100_C8VIC5 Phosphatidylserine decarboxylase Psd2, putative... 55 5e-06
UniRef100_Q62769-2 Isoform 2 of Protein unc-13 homolog B n=1 Tax... 55 5e-06
UniRef100_Q62769 Protein unc-13 homolog B n=1 Tax=Rattus norvegi... 55 5e-06
UniRef100_Q9Z1N9 Protein unc-13 homolog B n=1 Tax=Mus musculus R... 55 5e-06
UniRef100_P29101 Synaptotagmin-2 n=1 Tax=Rattus norvegicus RepID... 55 5e-06
UniRef100_P46097 Synaptotagmin-2 n=1 Tax=Mus musculus RepID=SYT2... 55 5e-06
UniRef100_Q8N9I0 Synaptotagmin-2 n=1 Tax=Homo sapiens RepID=SYT2... 55 5e-06
UniRef100_Q9BSJ8-2 Isoform 2 of Extended synaptotagmin-1 n=1 Tax... 55 5e-06
UniRef100_Q9BSJ8 Extended synaptotagmin-1 n=1 Tax=Homo sapiens R... 55 5e-06
UniRef100_UPI0001791492 PREDICTED: similar to CG6643 CG6643-PA n... 54 7e-06
UniRef100_UPI00015B4FEC PREDICTED: similar to CG15078-PA n=1 Tax... 54 7e-06
UniRef100_UPI00006A063A Synaptotagmin-5 (Synaptotagmin V) (SytV)... 54 7e-06
UniRef100_UPI00017B2FD4 UPI00017B2FD4 related cluster n=1 Tax=Te... 54 7e-06
UniRef100_UPI00017B29A7 UPI00017B29A7 related cluster n=1 Tax=Te... 54 7e-06
UniRef100_Q4S5D6 Chromosome 19 SCAF14731, whole genome shotgun s... 54 7e-06
UniRef100_UPI00016E29D5 UPI00016E29D5 related cluster n=1 Tax=Ta... 54 7e-06
UniRef100_UPI00016E29B6 UPI00016E29B6 related cluster n=1 Tax=Ta... 54 7e-06
UniRef100_Q4SL13 Chromosome 17 SCAF14563, whole genome shotgun s... 54 7e-06
UniRef100_C0JAE9 ZAC (Fragment) n=1 Tax=Oryza granulata RepID=C0... 54 7e-06
UniRef100_B4FFK7 GTPase activating protein n=1 Tax=Zea mays RepI... 54 7e-06
UniRef100_A9TQ09 Plant synaptotagmin: integral membrane double C... 54 7e-06
UniRef100_Q9V483 Unc-13, isoform C n=1 Tax=Drosophila melanogast... 54 7e-06
UniRef100_Q8IM86 Unc-13, isoform B n=1 Tax=Drosophila melanogast... 54 7e-06
UniRef100_C4QLP9 Unc-13 (Munc13), putative (Fragment) n=1 Tax=Sc... 54 7e-06
UniRef100_C4Q3D3 Unc-13 (Munc13), putative n=1 Tax=Schistosoma m... 54 7e-06
UniRef100_B4R2H5 Unc-13 n=1 Tax=Drosophila simulans RepID=B4R2H5... 54 7e-06
UniRef100_B4JZV1 GH24009 n=1 Tax=Drosophila grimshawi RepID=B4JZ... 54 7e-06
UniRef100_B4HC83 GL15609 n=1 Tax=Drosophila persimilis RepID=B4H... 54 7e-06
UniRef100_B0WHF9 Phorbol ester/diacylglycerol-binding protein un... 54 7e-06
UniRef100_UPI00017920D2 PREDICTED: similar to unc-13 CG2999-PB n... 54 9e-06
UniRef100_UPI0000E7FCD6 PREDICTED: similar to KIAA1228 protein n... 54 9e-06
UniRef100_UPI00006A4901 PREDICTED: similar to unc-13 homolog A n... 54 9e-06
UniRef100_UPI00016E9FE7 UPI00016E9FE7 related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FCB UPI00016E9FCB related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FCA UPI00016E9FCA related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FC9 UPI00016E9FC9 related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FC8 UPI00016E9FC8 related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FB2 UPI00016E9FB2 related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI00016E9FB1 UPI00016E9FB1 related cluster n=1 Tax=Ta... 54 9e-06
UniRef100_UPI0000ECBF69 Unc-13 homolog B (Munc13-2) (munc13). n=... 54 9e-06
UniRef100_UPI0000ECBF68 Unc-13 homolog B (Munc13-2) (munc13). n=... 54 9e-06
UniRef100_C0JA18 ZAC n=2 Tax=Oryza RepID=C0JA18_ORYGL 54 9e-06
UniRef100_C0J9Y5 ZAC n=2 Tax=Oryza RepID=C0J9Y5_ORYNI 54 9e-06
UniRef100_B9FU38 Putative uncharacterized protein n=1 Tax=Oryza ... 54 9e-06
UniRef100_B8LL63 Putative uncharacterized protein n=1 Tax=Picea ... 54 9e-06
UniRef100_B8B4J9 Putative uncharacterized protein n=1 Tax=Oryza ... 54 9e-06
UniRef100_A9PCD0 Putative uncharacterized protein n=1 Tax=Populu... 54 9e-06
UniRef100_Q17IK9 Putative uncharacterized protein n=1 Tax=Aedes ... 54 9e-06
UniRef100_A7RZG6 Predicted protein (Fragment) n=1 Tax=Nematostel... 54 9e-06
UniRef100_B6H2R6 Pc13g15440 protein n=1 Tax=Penicillium chrysoge... 54 9e-06
UniRef100_B2W6L2 Phosphatidylserine decarboxylase proenzyme n=1 ... 54 9e-06
UniRef100_P24505 Synaptotagmin-A n=1 Tax=Discopyge ommata RepID=... 54 9e-06