[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
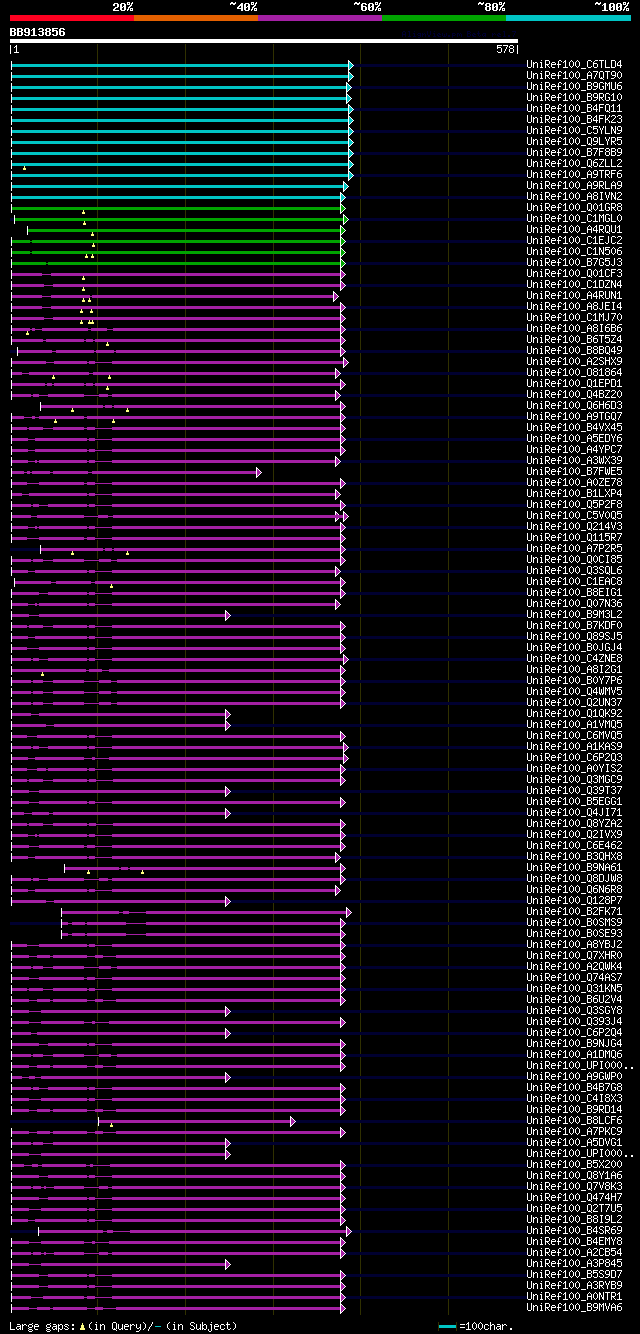
Query= BB913856 RCE13318
(578 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_C6TLD4 Putative uncharacterized protein n=1 Tax=Glycin... 255 2e-66
UniRef100_A7QT90 Chromosome chr1 scaffold_166, whole genome shot... 253 7e-66
UniRef100_B9GMU6 Predicted protein (Fragment) n=1 Tax=Populus tr... 252 1e-65
UniRef100_B9RG10 Fk506-binding protein, putative n=1 Tax=Ricinus... 248 3e-64
UniRef100_B4FQ11 Putative uncharacterized protein n=1 Tax=Zea ma... 247 5e-64
UniRef100_B4FK23 FK506 binding protein n=1 Tax=Zea mays RepID=B4... 247 5e-64
UniRef100_C5YLN9 Putative uncharacterized protein Sb07g021890 n=... 244 3e-63
UniRef100_Q9LYR5 Probable FKBP-type peptidyl-prolyl cis-trans is... 244 5e-63
UniRef100_B7F8B9 Putative uncharacterized protein n=1 Tax=Oryza ... 243 1e-62
UniRef100_Q6ZLL2 Immunophilin/FKBP-type peptidyl-prolyl cis-tran... 230 5e-59
UniRef100_A9TRF6 Predicted protein (Fragment) n=1 Tax=Physcomitr... 223 1e-56
UniRef100_A9RLA9 Predicted protein (Fragment) n=1 Tax=Physcomitr... 221 3e-56
UniRef100_A8IVN2 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 191 5e-47
UniRef100_Q01GR8 FKBP-type peptidyl-prolyl cis-trans isomerase (... 169 1e-40
UniRef100_C1MGL0 Predicted protein n=1 Tax=Micromonas pusilla CC... 166 1e-39
UniRef100_A4RQU1 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 164 6e-39
UniRef100_C1EJC2 Predicted protein n=1 Tax=Micromonas sp. RCC299... 117 5e-25
UniRef100_C1N506 Predicted protein (Fragment) n=1 Tax=Micromonas... 115 3e-24
UniRef100_B7G5J3 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 112 2e-23
UniRef100_Q01CF3 FKBP-type peptidyl-prolyl cis-trans isomerase (... 103 7e-21
UniRef100_C1DZN4 Predicted protein n=1 Tax=Micromonas sp. RCC299... 103 7e-21
UniRef100_A4RUN1 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 103 1e-20
UniRef100_A8JEI4 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 87 7e-16
UniRef100_C1MJ70 Predicted protein n=1 Tax=Micromonas pusilla CC... 87 9e-16
UniRef100_A8I6B6 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 65 4e-09
UniRef100_B6T5Z4 FK506 binding protein n=1 Tax=Zea mays RepID=B6... 65 5e-09
UniRef100_B8BQ49 Predicted protein n=1 Tax=Thalassiosira pseudon... 64 8e-09
UniRef100_A2SHX9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Met... 63 1e-08
UniRef100_O81864 Putative uncharacterized protein AT4g19830 n=1 ... 63 1e-08
UniRef100_Q1EPD1 Peptidylprolyl isomerase, FKBP-type family prot... 63 1e-08
UniRef100_Q4BZ20 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cro... 63 2e-08
UniRef100_Q6H6D3 Os02g0168700 protein n=2 Tax=Oryza sativa RepID... 62 2e-08
UniRef100_A9TGQ7 Predicted protein (Fragment) n=1 Tax=Physcomitr... 62 2e-08
UniRef100_B4VX45 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Mic... 62 3e-08
UniRef100_A5EDY6 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bra... 62 4e-08
UniRef100_A4YPC7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bra... 62 4e-08
UniRef100_A3WX39 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Nit... 61 7e-08
UniRef100_B7FWE5 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 61 7e-08
UniRef100_A0ZE78 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Nod... 60 9e-08
UniRef100_B1LXP4 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Met... 60 1e-07
UniRef100_Q5P2F8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Aro... 60 2e-07
UniRef100_C5V0Q5 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Gal... 60 2e-07
UniRef100_Q214V3 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Rho... 59 2e-07
UniRef100_Q115R7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Tri... 59 2e-07
UniRef100_A7P2R5 Chromosome chr1 scaffold_5, whole genome shotgu... 59 2e-07
UniRef100_Q0CI85 FK506-binding protein 1A n=1 Tax=Aspergillus te... 59 2e-07
UniRef100_Q3SQL6 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Nit... 59 3e-07
UniRef100_C1EAC8 Predicted protein n=1 Tax=Micromonas sp. RCC299... 59 3e-07
UniRef100_B8EIG1 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Met... 59 4e-07
UniRef100_Q07N36 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Rho... 58 5e-07
UniRef100_B9M3L2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 58 5e-07
UniRef100_B7KDF0 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 58 5e-07
UniRef100_Q89SJ5 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bra... 58 6e-07
UniRef100_B0JGJ4 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Mic... 58 6e-07
UniRef100_C4ZNE8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Tha... 58 6e-07
UniRef100_A8I2G1 Peptidyl-prolyl cis-trans isomerase, FKBP-type ... 58 6e-07
UniRef100_B0Y7P6 FKBP-type peptidyl-prolyl isomerase, putative n... 58 6e-07
UniRef100_Q4WMV5 FK506-binding protein 4 n=2 Tax=Aspergillus fum... 58 6e-07
UniRef100_Q2UN37 FK506-binding protein 4 n=2 Tax=Aspergillus Rep... 57 8e-07
UniRef100_Q1QK92 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Nit... 57 1e-06
UniRef100_A1VMQ5 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pol... 57 1e-06
UniRef100_C6MVQ5 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 57 1e-06
UniRef100_A1KAS9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Azo... 57 1e-06
UniRef100_C6P2Q3 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Sid... 57 1e-06
UniRef100_A0YIS2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Lyn... 57 1e-06
UniRef100_Q3MGC9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ana... 56 2e-06
UniRef100_Q39T37 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 56 2e-06
UniRef100_B5EGG1 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 56 2e-06
UniRef100_Q4JI71 45 kDa immunophilin FKBP45 n=1 Tax=Bombyx mori ... 56 2e-06
UniRef100_Q8YZA2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Nos... 56 2e-06
UniRef100_Q2IVX9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Rho... 56 2e-06
UniRef100_C6E462 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 56 2e-06
UniRef100_B3QHX8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Rho... 56 2e-06
UniRef100_B9NA61 Predicted protein n=1 Tax=Populus trichocarpa R... 56 2e-06
UniRef100_Q8DJW8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=The... 55 3e-06
UniRef100_Q6N6R8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Rho... 55 3e-06
UniRef100_Q128P7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pol... 55 3e-06
UniRef100_B2FK71 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ste... 55 3e-06
UniRef100_B0SMS9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Lep... 55 3e-06
UniRef100_B0SE93 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Lep... 55 3e-06
UniRef100_A8YBJ2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Mic... 55 3e-06
UniRef100_Q7XHR0 Os07g0490400 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_A2QWK4 Contig An11c0220, complete genome n=1 Tax=Asper... 55 3e-06
UniRef100_Q74AS7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Geo... 55 4e-06
UniRef100_Q31KN5 Peptidyl-prolyl cis-trans isomerase n=2 Tax=Syn... 55 4e-06
UniRef100_B6U2V4 FK506 binding protein n=1 Tax=Zea mays RepID=B6... 55 4e-06
UniRef100_Q3SGY8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Thi... 55 5e-06
UniRef100_Q393J4 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bur... 55 5e-06
UniRef100_C6P2Q4 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Sid... 55 5e-06
UniRef100_B9NJG4 Predicted protein n=2 Tax=cellular organisms Re... 55 5e-06
UniRef100_A1DMQ6 FKBP-type peptidyl-prolyl isomerase, putative n... 55 5e-06
UniRef100_UPI0001984CEE PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_A9GWP0 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Sor... 54 7e-06
UniRef100_B4B7G8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 54 7e-06
UniRef100_C4I8X3 Peptidyl-prolyl cis-trans isomerase n=12 Tax=Bu... 54 7e-06
UniRef100_B9RD14 Fk506-binding protein, putative n=1 Tax=Ricinus... 54 7e-06
UniRef100_B8LCF6 Predicted protein n=1 Tax=Thalassiosira pseudon... 54 7e-06
UniRef100_A7PKC9 Chromosome chr15 scaffold_19, whole genome shot... 54 7e-06
UniRef100_A5DVG1 Putative uncharacterized protein n=1 Tax=Lodder... 54 7e-06
UniRef100_UPI00016A9A6C FK506-binding protein n=1 Tax=Burkholder... 54 9e-06
UniRef100_B5X200 FK506-binding protein 1A n=1 Tax=Salmo salar Re... 54 9e-06
UniRef100_Q8Y1A6 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ral... 54 9e-06
UniRef100_Q7V8K3 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pro... 54 9e-06
UniRef100_Q474H7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ral... 54 9e-06
UniRef100_Q2T7U5 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bur... 54 9e-06
UniRef100_B8I9L2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Met... 54 9e-06
UniRef100_B4SR69 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ste... 54 9e-06
UniRef100_B4EMY8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Bur... 54 9e-06
UniRef100_A2CB54 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pro... 54 9e-06
UniRef100_A3P845 Peptidyl-prolyl cis-trans isomerase n=2 Tax=Bur... 54 9e-06
UniRef100_B5S9D7 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Ral... 54 9e-06
UniRef100_A3RYB9 Peptidyl-prolyl cis-trans isomerase n=2 Tax=Ral... 54 9e-06
UniRef100_A0NTR1 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Lab... 54 9e-06
UniRef100_B9MVA6 Predicted protein n=1 Tax=Populus trichocarpa R... 54 9e-06
UniRef100_C7YKQ8 Putative uncharacterized protein n=1 Tax=Nectri... 54 9e-06