[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB911043 RCE10092
(578 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
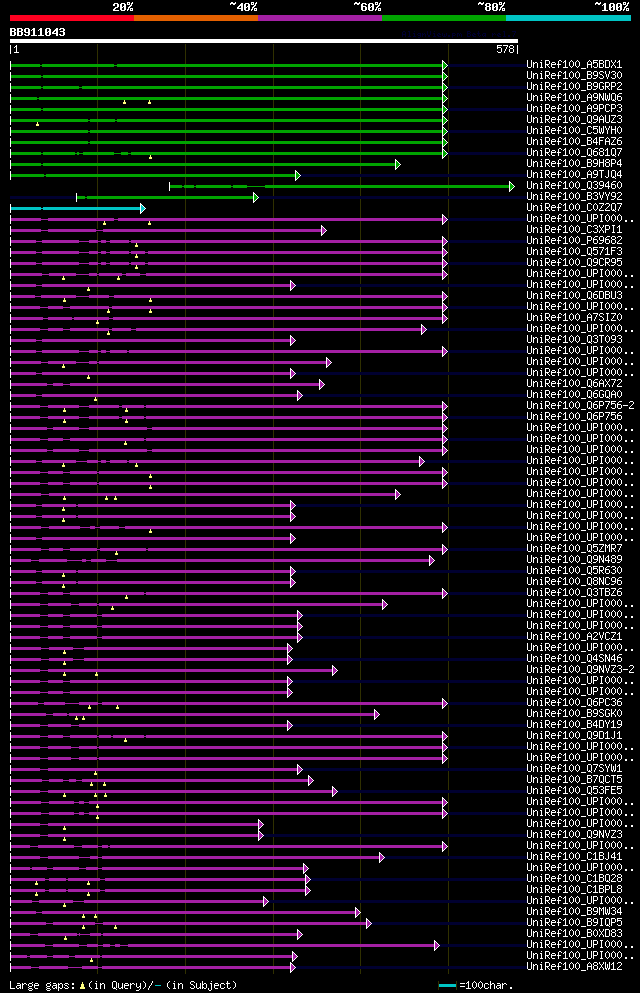
significant alignments:[graphical|details]
UniRef100_A5BDX1 Chromosome chr7 scaffold_44, whole genome shotg... 217 5e-55
UniRef100_B9SV30 Adaptin ear-binding coat-associated protein, pu... 198 3e-49
UniRef100_B9GRP2 ABC transporter family protein n=1 Tax=Populus ... 196 1e-48
UniRef100_A9NWQ6 Putative uncharacterized protein n=1 Tax=Picea ... 196 1e-48
UniRef100_A9PCP3 Putative uncharacterized protein n=1 Tax=Populu... 195 2e-48
UniRef100_Q9AUZ3 Os10g0476000 protein n=3 Tax=Oryza sativa RepID... 186 1e-45
UniRef100_C5WYH0 Putative uncharacterized protein Sb01g019420 n=... 184 4e-45
UniRef100_B4FAZ6 Adaptin ear-binding coat-associated protein 2 n... 184 6e-45
UniRef100_Q681Q7 Uncharacterized protein At1g03900 n=1 Tax=Arabi... 180 8e-44
UniRef100_B9H8P4 ABC transporter family protein n=1 Tax=Populus ... 177 5e-43
UniRef100_A9TJQ4 Predicted protein (Fragment) n=1 Tax=Physcomitr... 112 3e-23
UniRef100_Q39460 C.arietinum open reading frame n=1 Tax=Cicer ar... 107 5e-22
UniRef100_B3VY92 ABC transporter family protein (Fragment) n=1 T... 86 3e-15
UniRef100_C0Z2Q7 AT1G03900 protein n=1 Tax=Arabidopsis thaliana ... 82 2e-14
UniRef100_UPI000186ABEA hypothetical protein BRAFLDRAFT_110387 n... 68 6e-10
UniRef100_C3XPI1 Putative uncharacterized protein n=1 Tax=Branch... 66 2e-09
UniRef100_P69682 Adaptin ear-binding coat-associated protein 1 n... 66 2e-09
UniRef100_Q571F3 MFLJ00061 protein (Fragment) n=1 Tax=Mus muscul... 65 3e-09
UniRef100_Q9CR95 Adaptin ear-binding coat-associated protein 1 n... 65 3e-09
UniRef100_UPI00017B256D UPI00017B256D related cluster n=1 Tax=Te... 65 5e-09
UniRef100_UPI000194B9AC PREDICTED: similar to adaptin ear-bindin... 64 1e-08
UniRef100_Q6DBU3 LOC553536 protein (Fragment) n=1 Tax=Danio reri... 63 1e-08
UniRef100_UPI0000EB3BF7 Adaptin ear-binding coat-associated prot... 62 2e-08
UniRef100_A7SIZ0 Predicted protein n=1 Tax=Nematostella vectensi... 62 2e-08
UniRef100_UPI0000EB3BF6 Adaptin ear-binding coat-associated prot... 62 4e-08
UniRef100_Q3T093 Adaptin ear-binding coat-associated protein 1 n... 62 4e-08
UniRef100_UPI000155E5AC PREDICTED: similar to NECAP endocytosis ... 61 5e-08
UniRef100_UPI00016E58A9 UPI00016E58A9 related cluster n=1 Tax=Ta... 61 5e-08
UniRef100_UPI0000ECD52A PREDICTED: Gallus gallus hypothetical pr... 61 5e-08
UniRef100_Q6AX72 MGC83534 protein n=1 Tax=Xenopus laevis RepID=Q... 61 5e-08
UniRef100_Q6GQA0 MGC80231 protein n=1 Tax=Xenopus laevis RepID=Q... 61 7e-08
UniRef100_Q6P756-2 Isoform 2 of Adaptin ear-binding coat-associa... 61 7e-08
UniRef100_Q6P756 Adaptin ear-binding coat-associated protein 2 n... 61 7e-08
UniRef100_UPI000194DA13 PREDICTED: similar to adaptin-ear-bindin... 60 1e-07
UniRef100_UPI0001795C25 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI000194DA12 PREDICTED: adaptin-ear-binding coat-asso... 60 2e-07
UniRef100_UPI00006D1CA2 PREDICTED: similar to adaptin-ear-bindin... 60 2e-07
UniRef100_UPI00005A03CE PREDICTED: similar to Adaptin ear-bindin... 60 2e-07
UniRef100_UPI00004BD43C PREDICTED: similar to Adaptin ear-bindin... 60 2e-07
UniRef100_UPI0000ECA28A adaptin-ear-binding coat-associated prot... 60 2e-07
UniRef100_UPI0000E22FEE PREDICTED: hypothetical protein isoform ... 59 2e-07
UniRef100_UPI0000368847 PREDICTED: hypothetical protein isoform ... 59 2e-07
UniRef100_UPI00016E2D7A UPI00016E2D7A related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_UPI00004BE9AB PREDICTED: similar to adaptin-ear-bindin... 59 2e-07
UniRef100_Q5ZMR7 Putative uncharacterized protein n=1 Tax=Gallus... 59 2e-07
UniRef100_Q9N489 Putative uncharacterized protein n=1 Tax=Caenor... 59 2e-07
UniRef100_Q5R630 Adaptin ear-binding coat-associated protein 1 n... 59 2e-07
UniRef100_Q8NC96 Adaptin ear-binding coat-associated protein 1 n... 59 2e-07
UniRef100_Q3TBZ6 Putative uncharacterized protein n=1 Tax=Mus mu... 59 3e-07
UniRef100_UPI0000367EA3 PREDICTED: hypothetical protein isoform ... 59 4e-07
UniRef100_UPI00006A1C9E Adaptin ear-binding coat-associated prot... 59 4e-07
UniRef100_UPI00006A1C9D Adaptin ear-binding coat-associated prot... 59 4e-07
UniRef100_A2VCZ1 LOC100037850 protein n=1 Tax=Xenopus (Silurana)... 59 4e-07
UniRef100_UPI00017B1A49 UPI00017B1A49 related cluster n=1 Tax=Te... 58 5e-07
UniRef100_Q4SN46 Chromosome 8 SCAF14543, whole genome shotgun se... 58 5e-07
UniRef100_Q9NVZ3-2 Isoform 2 of Adaptin ear-binding coat-associa... 58 6e-07
UniRef100_UPI0000D9975B PREDICTED: NECAP endocytosis associated ... 57 1e-06
UniRef100_UPI00006D647F PREDICTED: NECAP endocytosis associated ... 57 1e-06
UniRef100_Q6PC36 Novel protein similar to vertebrate NECAP endoc... 57 1e-06
UniRef100_B9SGK0 Adaptin ear-binding coat-associated protein, pu... 57 1e-06
UniRef100_B4DY19 cDNA FLJ53612, highly similar to Adaptin ear-bi... 57 1e-06
UniRef100_Q9D1J1 Adaptin ear-binding coat-associated protein 2 n... 57 1e-06
UniRef100_UPI0000E1E688 PREDICTED: hypothetical protein isoform ... 57 1e-06
UniRef100_UPI0000367EA2 PREDICTED: hypothetical protein isoform ... 57 1e-06
UniRef100_Q7SYW1 MGC64450 protein n=1 Tax=Xenopus laevis RepID=Q... 57 1e-06
UniRef100_B7QCT5 Adaptin ear-binding coat-associated protein, pu... 57 1e-06
UniRef100_Q53FE5 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_UPI0000E4A336 PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000E4600E PREDICTED: hypothetical protein, partial... 56 2e-06
UniRef100_UPI0001948488 NECAP endocytosis associated 2 isoform 3... 56 2e-06
UniRef100_Q9NVZ3 Adaptin ear-binding coat-associated protein 2 n... 56 2e-06
UniRef100_UPI00015B4FB3 PREDICTED: similar to CG9132-PB n=1 Tax=... 56 2e-06
UniRef100_C1BJ41 Adaptin ear-binding coat-associated protein 1 n... 56 2e-06
UniRef100_UPI0000516D9C PREDICTED: similar to CG9132-PB, isoform... 55 3e-06
UniRef100_C1BQ28 NECAP-like protein CG9132 n=1 Tax=Caligus roger... 55 3e-06
UniRef100_C1BPL8 NECAP-like protein CG9132 n=1 Tax=Caligus roger... 55 3e-06
UniRef100_UPI00016E68EA UPI00016E68EA related cluster n=1 Tax=Ta... 55 4e-06
UniRef100_B9MW34 Predicted protein n=1 Tax=Populus trichocarpa R... 55 5e-06
UniRef100_B9IQP5 Predicted protein n=1 Tax=Populus trichocarpa R... 55 5e-06
UniRef100_B0XD83 Adaptin ear-binding coat-associated protein 2 n... 55 5e-06
UniRef100_UPI000186F26D conserved hypothetical protein n=1 Tax=P... 54 7e-06
UniRef100_UPI00017938B4 PREDICTED: similar to adaptin ear-bindin... 54 7e-06
UniRef100_A8XW12 C. briggsae CBR-UBC-15 protein n=1 Tax=Caenorha... 54 7e-06
UniRef100_B2WG42 Adaptin ear-binding coat-associated protein 2 n... 54 9e-06