[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB909591 RCE08433
(577 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
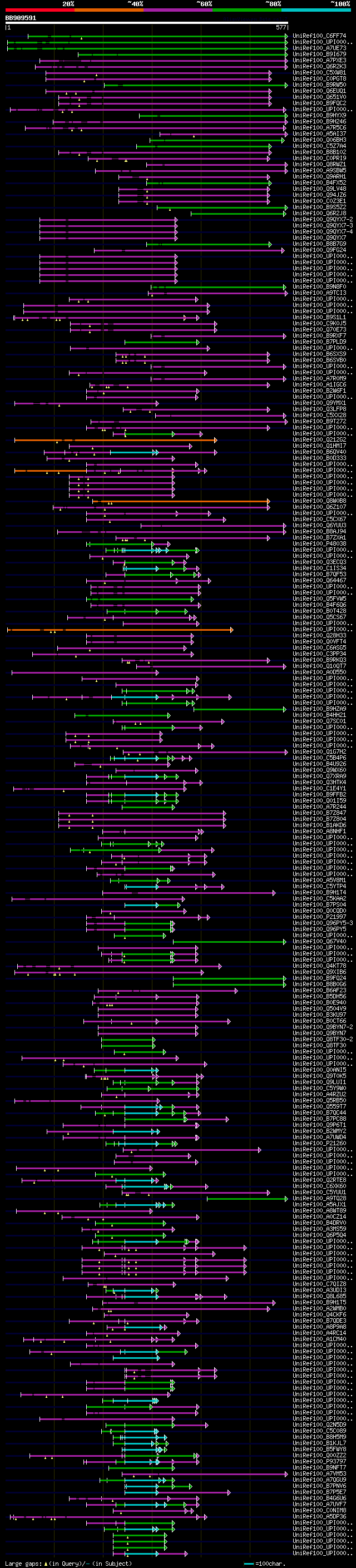
significant alignments:[graphical|details]
UniRef100_C6FF74 LRR receptor-like kinase n=1 Tax=Glycine max Re... 244 5e-63
UniRef100_UPI0001984556 PREDICTED: hypothetical protein n=1 Tax=... 176 2e-42
UniRef100_A7UE73 LRR receptor-like kinase n=1 Tax=Solanum tubero... 166 9e-40
UniRef100_B9I679 Predicted protein (Fragment) n=1 Tax=Populus tr... 139 2e-31
UniRef100_A7PXE3 Chromosome chr12 scaffold_36, whole genome shot... 122 3e-26
UniRef100_Q6R2K3 Protein STRUBBELIG-RECEPTOR FAMILY 3 n=2 Tax=Ar... 121 4e-26
UniRef100_C5XW81 Putative uncharacterized protein Sb04g005115 n=... 119 2e-25
UniRef100_C0PGT8 Putative uncharacterized protein n=1 Tax=Zea ma... 117 5e-25
UniRef100_B9RW50 Lrr receptor-linked protein kinase, putative n=... 116 1e-24
UniRef100_Q6EUQ1 Os02g0176100 protein n=1 Tax=Oryza sativa Japon... 113 9e-24
UniRef100_Q651V0 Putative leucine-rich repeat transmembrane prot... 100 1e-19
UniRef100_B9FQC2 Putative uncharacterized protein n=1 Tax=Oryza ... 100 1e-19
UniRef100_UPI0001985F91 PREDICTED: hypothetical protein n=1 Tax=... 96 3e-18
UniRef100_B9HYX9 Predicted protein (Fragment) n=1 Tax=Populus tr... 96 3e-18
UniRef100_B9H246 Predicted protein n=1 Tax=Populus trichocarpa R... 95 3e-18
UniRef100_A7R5C6 Chromosome undetermined scaffold_937, whole gen... 95 3e-18
UniRef100_A5AI37 Putative uncharacterized protein n=1 Tax=Vitis ... 94 6e-18
UniRef100_Q06BH3 Protein STRUBBELIG-RECEPTOR FAMILY 1 n=2 Tax=Ar... 92 3e-17
UniRef100_C5Z7A4 Putative uncharacterized protein Sb10g026340 n=... 92 4e-17
UniRef100_B8B102 Putative uncharacterized protein n=1 Tax=Oryza ... 89 3e-16
UniRef100_C0PRI9 Putative uncharacterized protein n=1 Tax=Picea ... 78 6e-13
UniRef100_Q8RWZ1 Protein STRUBBELIG n=2 Tax=Arabidopsis thaliana... 76 2e-12
UniRef100_A9SBW5 Predicted protein (Fragment) n=1 Tax=Physcomitr... 75 3e-12
UniRef100_Q9ARH1 Receptor protein kinase PERK1 n=1 Tax=Brassica ... 74 6e-12
UniRef100_B4FX52 Putative uncharacterized protein n=1 Tax=Zea ma... 74 8e-12
UniRef100_Q9LV48 Protein kinase-like protein n=1 Tax=Arabidopsis... 73 2e-11
UniRef100_Q94JZ6 At3g24600 n=1 Tax=Arabidopsis thaliana RepID=Q9... 73 2e-11
UniRef100_C0Z3E1 AT3G24550 protein n=1 Tax=Arabidopsis thaliana ... 73 2e-11
UniRef100_B9S5Z2 Lrr receptor-linked protein kinase, putative n=... 73 2e-11
UniRef100_Q6R2J8 Protein STRUBBELIG-RECEPTOR FAMILY 8 n=3 Tax=Ar... 73 2e-11
UniRef100_Q9QYX7-2 Isoform 2 of Protein piccolo n=1 Tax=Mus musc... 72 3e-11
UniRef100_Q9QYX7-3 Isoform 3 of Protein piccolo n=1 Tax=Mus musc... 72 3e-11
UniRef100_Q9QYX7-4 Isoform 4 of Protein piccolo n=1 Tax=Mus musc... 72 3e-11
UniRef100_Q9QYX7 Protein piccolo n=1 Tax=Mus musculus RepID=PCLO... 72 3e-11
UniRef100_B8B7G9 Putative uncharacterized protein n=1 Tax=Oryza ... 72 4e-11
UniRef100_Q9FG24 Protein STRUBBELIG-RECEPTOR FAMILY 2 n=2 Tax=Ar... 71 5e-11
UniRef100_UPI00015FF5B0 piccolo isoform 2 n=1 Tax=Mus musculus R... 70 9e-11
UniRef100_UPI00015FA08D piccolo isoform 1 n=1 Tax=Mus musculus R... 70 9e-11
UniRef100_UPI00016D3858 piccolo (presynaptic cytomatrix protein)... 70 9e-11
UniRef100_UPI00016D3827 piccolo (presynaptic cytomatrix protein)... 70 9e-11
UniRef100_UPI00015DF6C1 piccolo (presynaptic cytomatrix protein)... 70 9e-11
UniRef100_B9N8F0 Predicted protein n=1 Tax=Populus trichocarpa R... 70 9e-11
UniRef100_A9TCI3 Predicted protein n=1 Tax=Physcomitrella patens... 70 1e-10
UniRef100_UPI00005A3937 PREDICTED: similar to formin-like 2 isof... 70 2e-10
UniRef100_UPI0001B7C139 Protein piccolo (Aczonin) (Multidomain p... 70 2e-10
UniRef100_UPI0001B7C138 Protein piccolo (Aczonin) (Multidomain p... 70 2e-10
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 70 2e-10
UniRef100_C9K0J5 Putative uncharacterized protein RAPH1 n=1 Tax=... 70 2e-10
UniRef100_Q70E73 Ras-associated and pleckstrin homology domains-... 70 2e-10
UniRef100_B9RXF7 Serine/threonine-protein kinase PBS1, putative ... 69 2e-10
UniRef100_B7PLD9 Circumsporozoite protein, putative (Fragment) n... 69 2e-10
UniRef100_UPI0001B7C13A Protein piccolo (Aczonin) (Multidomain p... 69 3e-10
UniRef100_B6SXS9 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 69 3e-10
UniRef100_B6SVB0 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 69 3e-10
UniRef100_UPI0001984178 PREDICTED: hypothetical protein n=1 Tax=... 69 3e-10
UniRef100_UPI000069FBE2 Formin-like protein 2 (Formin homology 2... 69 3e-10
UniRef100_A7R0M9 Chromosome chr10 scaffold_312, whole genome sho... 69 3e-10
UniRef100_A1IGC6 PERK1-like protein kinase n=1 Tax=Nicotiana tab... 69 3e-10
UniRef100_B2W6F1 Putative uncharacterized protein n=1 Tax=Pyreno... 68 4e-10
UniRef100_UPI0001951250 Formin-like protein 2 (Formin homology 2... 68 6e-10
UniRef100_Q9YMX1 Essential structural protein pp78-81 n=1 Tax=Ly... 68 6e-10
UniRef100_Q3LFP8 PERK1-like protein kinase n=1 Tax=Glycine max R... 68 6e-10
UniRef100_C5XX28 Putative uncharacterized protein Sb04g006110 n=... 68 6e-10
UniRef100_B9T272 Receptor protein kinase CLAVATA1, putative n=1 ... 68 6e-10
UniRef100_UPI000198414A PREDICTED: hypothetical protein isoform ... 67 8e-10
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 67 8e-10
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 67 8e-10
UniRef100_Q1HMI7 Formin B n=2 Tax=Trypanosoma cruzi RepID=Q1HMI7... 67 8e-10
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 67 8e-10
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 67 8e-10
UniRef100_UPI00017F09BE PREDICTED: similar to KIAA1902 protein, ... 67 1e-09
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 67 1e-09
UniRef100_UPI0000E1F767 PREDICTED: formin-like 2 isoform 3 n=1 T... 67 1e-09
UniRef100_UPI0000E1F766 PREDICTED: formin-like 2 isoform 1 n=1 T... 67 1e-09
UniRef100_UPI0000E1F765 PREDICTED: formin-like 2 isoform 7 n=1 T... 67 1e-09
UniRef100_UPI0000E1F763 PREDICTED: formin-like 2 isoform 4 n=1 T... 67 1e-09
UniRef100_Q8W0B8 Os01g0227200 protein n=1 Tax=Oryza sativa Japon... 67 1e-09
UniRef100_Q6Z107 Os02g0549200 protein n=1 Tax=Oryza sativa Japon... 67 1e-09
UniRef100_UPI00000216EF glyceraldehyde-3-phosphate dehydrogenase... 67 1e-09
UniRef100_C5CX67 FHA domain containing protein n=1 Tax=Variovora... 67 1e-09
UniRef100_Q6YUU3 Os02g0190500 protein n=1 Tax=Oryza sativa Japon... 67 1e-09
UniRef100_B8AJ94 Putative uncharacterized protein n=1 Tax=Oryza ... 67 1e-09
UniRef100_B7ZXA1 Putative uncharacterized protein n=1 Tax=Zea ma... 67 1e-09
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 67 1e-09
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 66 2e-09
UniRef100_UPI0000F2B4E2 PREDICTED: similar to KIAA1100 protein n... 66 2e-09
UniRef100_Q3ECQ3 Uncharacterized protein At1g54215.1 n=1 Tax=Ara... 66 2e-09
UniRef100_C1IS34 Minicollagen-1 n=1 Tax=Malo kingi RepID=C1IS34_... 66 2e-09
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 66 2e-09
UniRef100_Q64467 Glyceraldehyde-3-phosphate dehydrogenase, testi... 66 2e-09
UniRef100_UPI0000F21440 PREDICTED: similar to RING finger protei... 66 2e-09
UniRef100_UPI0001A2BDC1 UPI0001A2BDC1 related cluster n=1 Tax=Da... 66 2e-09
UniRef100_Q5FVW5 LOC548379 protein (Fragment) n=1 Tax=Xenopus (S... 66 2e-09
UniRef100_B4F6Q6 Putative uncharacterized protein (Fragment) n=1... 66 2e-09
UniRef100_B0T428 Glycoside hydrolase family 16 n=1 Tax=Caulobact... 66 2e-09
UniRef100_Q5CS67 Signal peptide containing large protein with pr... 66 2e-09
UniRef100_UPI0001926BC7 PREDICTED: similar to mini-collagen isof... 65 3e-09
UniRef100_UPI00015B42DB PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_Q28H33 Novel protein n=1 Tax=Xenopus (Silurana) tropic... 65 3e-09
UniRef100_Q0VFT4 Putative uncharacterized protein (Fragment) n=2... 65 3e-09
UniRef100_C6ASG5 Putative uncharacterized protein n=1 Tax=Rhizob... 65 3e-09
UniRef100_C3PP34 Actin polymerization protein RickA n=1 Tax=Rick... 65 3e-09
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 65 3e-09
UniRef100_Q10QT7 Os03g0183800 protein n=3 Tax=Oryza sativa RepID... 65 3e-09
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 65 3e-09
UniRef100_UPI0001926BAB PREDICTED: similar to mini-collagen n=1 ... 65 4e-09
UniRef100_UPI00017974C7 PREDICTED: similar to KIAA1902 protein n... 65 4e-09
UniRef100_UPI0000E2427E PREDICTED: guanine nucleotide binding pr... 65 4e-09
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 65 4e-09
UniRef100_UPI0000D9F166 PREDICTED: similar to guanine nucleotide... 65 4e-09
UniRef100_B9HZA9 Predicted protein n=1 Tax=Populus trichocarpa R... 65 4e-09
UniRef100_B4HH21 GM26598 n=1 Tax=Drosophila sechellia RepID=B4HH... 65 4e-09
UniRef100_Q7SC01 Putative uncharacterized protein n=1 Tax=Neuros... 65 4e-09
UniRef100_UPI0001985FA6 PREDICTED: hypothetical protein n=1 Tax=... 65 5e-09
UniRef100_UPI0000F2C4EF PREDICTED: similar to junction-mediating... 65 5e-09
UniRef100_UPI0000F2C4EE PREDICTED: similar to junction-mediating... 65 5e-09
UniRef100_UPI0001951234 Ras-associated and pleckstrin homology d... 65 5e-09
UniRef100_Q1G7H2 Dishevelled-associated activator of morphogenes... 65 5e-09
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 65 5e-09
UniRef100_B4U926 Putative uncharacterized protein n=1 Tax=Hydrog... 65 5e-09
UniRef100_Q9WX60 Ccp protein n=1 Tax=Gluconacetobacter xylinus R... 65 5e-09
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 65 5e-09
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 65 5e-09
UniRef100_C1E4Y1 Putative uncharacterized protein n=1 Tax=Microm... 65 5e-09
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 65 5e-09
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 65 5e-09
UniRef100_A7R244 Chromosome undetermined scaffold_398, whole gen... 65 5e-09
UniRef100_B7Z847 cDNA FLJ52583 n=1 Tax=Homo sapiens RepID=B7Z847... 65 5e-09
UniRef100_B7Z804 cDNA FLJ61626 n=1 Tax=Homo sapiens RepID=B7Z804... 65 5e-09
UniRef100_B1AKD6 Asparagine-linked glycosylation 13 homolog (S. ... 65 5e-09
UniRef100_A8NHF1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 65 5e-09
UniRef100_UPI0001797788 PREDICTED: similar to Zinc finger protei... 64 6e-09
UniRef100_UPI0000E23E7D PREDICTED: WAS protein homology region 2... 64 6e-09
UniRef100_UPI0000603C6F PREDICTED: Ras association (RalGDS/AF-6)... 64 6e-09
UniRef100_UPI000069FBE3 Formin-like protein 2 (Formin homology 2... 64 6e-09
UniRef100_UPI00004D6F7F Formin-like protein 2 (Formin homology 2... 64 6e-09
UniRef100_UPI0001B798A6 UPI0001B798A6 related cluster n=1 Tax=Ho... 64 6e-09
UniRef100_UPI000184A345 DMRT-like family B with proline-rich C-t... 64 6e-09
UniRef100_A5V8M1 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 64 6e-09
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 64 6e-09
UniRef100_B9H1T4 Predicted protein n=1 Tax=Populus trichocarpa R... 64 6e-09
UniRef100_C5KAA2 Putative uncharacterized protein n=1 Tax=Perkin... 64 6e-09
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 64 6e-09
UniRef100_Q0CQD0 Predicted protein n=1 Tax=Aspergillus terreus N... 64 6e-09
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 64 6e-09
UniRef100_Q96PY5-3 Isoform 2 of Formin-like protein 2 n=1 Tax=Ho... 64 6e-09
UniRef100_Q96PY5 Formin-like protein 2 n=1 Tax=Homo sapiens RepI... 64 6e-09
UniRef100_UPI0001795F40 PREDICTED: leiomodin 2 (cardiac) n=1 Tax... 64 8e-09
UniRef100_Q67V40 cDNA clone:J013050P20, full insert sequence n=2... 64 8e-09
UniRef100_UPI0000D9C719 PREDICTED: similar to zinc finger protei... 64 8e-09
UniRef100_UPI000069FBE4 Formin-like protein 2 (Formin homology 2... 64 8e-09
UniRef100_UPI00004D6F7D Formin-like protein 2 (Formin homology 2... 64 8e-09
UniRef100_Q4KT78 ORF1629 n=1 Tax=Chrysodeixis chalcites nucleopo... 64 8e-09
UniRef100_Q9XIB6 F13F21.7 protein n=1 Tax=Arabidopsis thaliana R... 64 8e-09
UniRef100_B9FQ24 Putative uncharacterized protein n=1 Tax=Oryza ... 64 8e-09
UniRef100_B8B0G6 Putative uncharacterized protein n=1 Tax=Oryza ... 64 8e-09
UniRef100_B6AF23 Putative uncharacterized protein n=1 Tax=Crypto... 64 8e-09
UniRef100_B5DH56 GA25354 n=1 Tax=Drosophila pseudoobscura pseudo... 64 8e-09
UniRef100_B0E940 Formin 2,3 and collagen domain-containing prote... 64 8e-09
UniRef100_Q504V9 ZNF341 protein (Fragment) n=1 Tax=Homo sapiens ... 64 8e-09
UniRef100_B3KU97 cDNA FLJ39436 fis, clone PROST2004739, highly s... 64 8e-09
UniRef100_B0CT66 RhoA GTPase effector DIA/Diaphanous n=1 Tax=Lac... 64 8e-09
UniRef100_Q9BYN7-2 Isoform 2 of Zinc finger protein 341 n=1 Tax=... 64 8e-09
UniRef100_Q9BYN7 Zinc finger protein 341 n=1 Tax=Homo sapiens Re... 64 8e-09
UniRef100_Q8TF30-2 Isoform 2 of WASP homolog-associated protein ... 64 8e-09
UniRef100_Q8TF30 WASP homolog-associated protein with actin, mem... 64 8e-09
UniRef100_UPI0000F2E472 PREDICTED: hypothetical protein n=1 Tax=... 64 1e-08
UniRef100_UPI0000D9B853 PREDICTED: similar to Formin-1 isoform I... 64 1e-08
UniRef100_UPI0001A2D80C UPI0001A2D80C related cluster n=1 Tax=Da... 64 1e-08
UniRef100_Q0ANI5 OmpA/MotB domain protein n=1 Tax=Maricaulis mar... 64 1e-08
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 64 1e-08
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 64 1e-08
UniRef100_C5Y9W0 Putative uncharacterized protein Sb06g019060 n=... 64 1e-08
UniRef100_A4RZU2 Predicted protein n=1 Tax=Ostreococcus lucimari... 64 1e-08
UniRef100_Q5RB50 Putative uncharacterized protein DKFZp459N037 n... 64 1e-08
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 64 1e-08
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 64 1e-08
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 64 1e-08
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 64 1e-08
UniRef100_B2WMY2 Putative uncharacterized protein n=1 Tax=Pyreno... 64 1e-08
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 64 1e-08
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 64 1e-08
UniRef100_UPI0000E7FD76 PREDICTED: similar to SH3 domain binding... 63 1e-08
UniRef100_UPI0000E1F764 PREDICTED: formin-like 2 isoform 6 n=1 T... 63 1e-08
UniRef100_UPI00005EB969 PREDICTED: similar to SET binding protei... 63 1e-08
UniRef100_UPI0000122471 Hypothetical protein CBG02907 n=1 Tax=Ca... 63 1e-08
UniRef100_UPI0001AE70C7 UPI0001AE70C7 related cluster n=1 Tax=Ho... 63 1e-08
UniRef100_Q2RTE8 Putative uncharacterized protein n=1 Tax=Rhodos... 63 1e-08
UniRef100_C6XK60 OmpA/MotB domain protein n=1 Tax=Hirschia balti... 63 1e-08
UniRef100_C5YUU1 Putative uncharacterized protein Sb09g007240 n=... 63 1e-08
UniRef100_A9TQ28 Predicted protein (Fragment) n=1 Tax=Physcomitr... 63 1e-08
UniRef100_A5AJX1 Putative uncharacterized protein n=1 Tax=Vitis ... 63 1e-08
UniRef100_A8WT89 C. briggsae CBR-SHN-1 protein n=1 Tax=Caenorhab... 63 1e-08
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 63 1e-08
UniRef100_B4DRV0 cDNA FLJ58570, highly similar to Mus musculus l... 63 1e-08
UniRef100_A3MS59 Putative uncharacterized protein n=1 Tax=Pyroba... 63 1e-08
UniRef100_Q6P5Q4 Leiomodin-2 n=1 Tax=Homo sapiens RepID=LMOD2_HUMAN 63 1e-08
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 63 2e-08
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 63 2e-08
UniRef100_UPI0000DB6FB3 PREDICTED: similar to plexus CG4444-PA n... 63 2e-08
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 63 2e-08
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 63 2e-08
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 63 2e-08
UniRef100_UPI0000F3293E PREDICTED: Bos taurus similar to bromodo... 63 2e-08
UniRef100_C7QIZ8 Putative uncharacterized protein n=1 Tax=Catenu... 63 2e-08
UniRef100_A3UDI3 OmpA family protein n=1 Tax=Oceanicaulis alexan... 63 2e-08
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 63 2e-08
UniRef100_B9H1T5 Predicted protein n=1 Tax=Populus trichocarpa R... 63 2e-08
UniRef100_A2WMB0 Putative uncharacterized protein n=1 Tax=Oryza ... 63 2e-08
UniRef100_Q4CKF6 Dispersed gene family protein 1 (DGF-1), putati... 63 2e-08
UniRef100_B7QDE3 Putative uncharacterized protein (Fragment) n=1... 63 2e-08
UniRef100_A8P9A8 Putative uncharacterized protein n=1 Tax=Brugia... 63 2e-08
UniRef100_A4RC14 Predicted protein n=1 Tax=Magnaporthe grisea Re... 63 2e-08
UniRef100_A1CM40 Cytokinesis protein SepA/Bni1 n=1 Tax=Aspergill... 63 2e-08
UniRef100_UPI00017C2CD7 PREDICTED: similar to formin-like 2 n=1 ... 62 2e-08
UniRef100_UPI000155487E PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI0000F2E28C PREDICTED: similar to high-mobility grou... 62 2e-08
UniRef100_UPI0000E80902 PREDICTED: similar to RAPH1 protein n=1 ... 62 2e-08
UniRef100_UPI0000E1F8E7 PREDICTED: Ras association and pleckstri... 62 2e-08
UniRef100_UPI0000E1F8E6 PREDICTED: Ras association and pleckstri... 62 2e-08
UniRef100_UPI0000E1F761 PREDICTED: formin-like 2 isoform 2 n=1 T... 62 2e-08
UniRef100_UPI0000E1F760 PREDICTED: formin-like 2 isoform 8 n=1 T... 62 2e-08
UniRef100_UPI000069FC8F cardiomyopathy associated 3 isoform 1 n=... 62 2e-08
UniRef100_UPI0000DC0BBF UPI0000DC0BBF related cluster n=1 Tax=Ra... 62 2e-08
UniRef100_UPI00016E4781 UPI00016E4781 related cluster n=1 Tax=Ta... 62 2e-08
UniRef100_UPI000179F39E UPI000179F39E related cluster n=1 Tax=Bo... 62 2e-08
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 62 2e-08
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 62 2e-08
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 62 2e-08
UniRef100_B8H5M9 Cell surface antigen Sca2 n=2 Tax=Caulobacter v... 62 2e-08
UniRef100_B1KJL7 Putative lipoprotein n=1 Tax=Shewanella woodyi ... 62 2e-08
UniRef100_B5FWY8 OmpA family protein n=1 Tax=Riemerella anatipes... 62 2e-08
UniRef100_Q00ZZ2 Membrane coat complex Retromer, subunit VPS5/SN... 62 2e-08
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 62 2e-08
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 62 2e-08
UniRef100_A7VM53 Receptor-like kinase n=1 Tax=Closterium ehrenbe... 62 2e-08
UniRef100_A7QGU9 Chromosome chr16 scaffold_94, whole genome shot... 62 2e-08
UniRef100_B7PNV6 Putative uncharacterized protein (Fragment) n=1... 62 2e-08
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 62 2e-08
UniRef100_B4G6U6 GL19111 n=1 Tax=Drosophila persimilis RepID=B4G... 62 2e-08
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 62 2e-08
UniRef100_C0NIM8 Putative uncharacterized protein n=1 Tax=Ajello... 62 2e-08
UniRef100_A5DP36 Actin cytoskeleton-regulatory complex protein P... 62 2e-08
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 62 3e-08
UniRef100_UPI0001553749 PREDICTED: hypothetical protein n=1 Tax=... 62 3e-08
UniRef100_UPI0000E254CE PREDICTED: piccolo isoform 3 n=1 Tax=Pan... 62 3e-08
UniRef100_UPI0000E254CD PREDICTED: piccolo isoform 2 n=1 Tax=Pan... 62 3e-08
UniRef100_UPI0000E254CC PREDICTED: piccolo isoform 1 n=1 Tax=Pan... 62 3e-08
UniRef100_UPI00005A6092 PREDICTED: similar to multidomain presyn... 62 3e-08
UniRef100_UPI00015DF6E5 enabled homolog (Drosophila) n=1 Tax=Mus... 62 3e-08
UniRef100_UPI0001573469 piccolo isoform 1 n=1 Tax=Homo sapiens R... 62 3e-08
UniRef100_UPI000156FA8C piccolo isoform 2 n=1 Tax=Homo sapiens R... 62 3e-08
UniRef100_UPI00016E7DED UPI00016E7DED related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E2CCB UPI00016E2CCB related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E2CCA UPI00016E2CCA related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI000179CD6A UPI000179CD6A related cluster n=1 Tax=Bo... 62 3e-08
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 62 3e-08
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 62 3e-08
UniRef100_Q2N609 Peptidoglycan-associated protein n=1 Tax=Erythr... 62 3e-08
UniRef100_B0T2W0 OmpA/MotB domain protein n=1 Tax=Caulobacter sp... 62 3e-08
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 62 3e-08
UniRef100_A5V4K4 OmpA/MotB domain protein n=1 Tax=Sphingomonas w... 62 3e-08
UniRef100_A5E9I8 Putative Peptidase, Caspase-like domain and TPR... 62 3e-08
UniRef100_B4W5S8 OmpA family protein n=1 Tax=Brevundimonas sp. B... 62 3e-08
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 62 3e-08
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 62 3e-08
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 62 3e-08
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 62 3e-08
UniRef100_B4LJT2 GJ20767 n=1 Tax=Drosophila virilis RepID=B4LJT2... 62 3e-08
UniRef100_B3M020 GF18870 n=1 Tax=Drosophila ananassae RepID=B3M0... 62 3e-08
UniRef100_A6NG74 Putative uncharacterized protein PCLO n=2 Tax=H... 62 3e-08
UniRef100_Q2H4B7 Predicted protein n=1 Tax=Chaetomium globosum R... 62 3e-08
UniRef100_Q9Y6V0-2 Isoform 2 of Protein piccolo n=1 Tax=Homo sap... 62 3e-08
UniRef100_Q9Y6V0 Protein piccolo n=1 Tax=Homo sapiens RepID=PCLO... 62 3e-08
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 62 3e-08
UniRef100_UPI00017C348A PREDICTED: zinc finger protein 341 n=1 T... 62 4e-08
UniRef100_UPI0000F2BFBF PREDICTED: similar to bromodomain PHD fi... 62 4e-08
UniRef100_UPI0000D9A916 PREDICTED: similar to leiomodin 2 (cardi... 62 4e-08
UniRef100_UPI0000D9A914 PREDICTED: similar to leiomodin 2 (cardi... 62 4e-08
UniRef100_UPI0000EE3DB8 zinc finger homeodomain 4 n=1 Tax=Homo s... 62 4e-08
UniRef100_UPI00004576BB Zinc finger homeobox protein 4 (Zinc fin... 62 4e-08
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 62 4e-08
UniRef100_UPI00016E98C1 UPI00016E98C1 related cluster n=1 Tax=Ta... 62 4e-08
UniRef100_UPI00016E98A2 UPI00016E98A2 related cluster n=1 Tax=Ta... 62 4e-08
UniRef100_UPI00016E5C42 UPI00016E5C42 related cluster n=1 Tax=Ta... 62 4e-08
UniRef100_UPI000195134F UPI000195134F related cluster n=1 Tax=Bo... 62 4e-08
UniRef100_Q1GRM3 OmpA/MotB n=1 Tax=Sphingopyxis alaskensis RepID... 62 4e-08
UniRef100_Q1RH03 Cell surface antigen Sca2 n=2 Tax=Rickettsia be... 62 4e-08
UniRef100_Q3L8Q3 Surface antigen (Fragment) n=1 Tax=Rickettsia b... 62 4e-08
UniRef100_C5JBL9 Uncharacterized protein n=1 Tax=uncultured bact... 62 4e-08
UniRef100_Q60E30 Putative uncharacterized protein OSJNBb0012L23.... 62 4e-08
UniRef100_B9FJ25 Putative uncharacterized protein n=1 Tax=Oryza ... 62 4e-08
UniRef100_A9TZ12 Predicted protein (Fragment) n=1 Tax=Physcomitr... 62 4e-08
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 62 4e-08
UniRef100_Q00484 Mini-collagen n=1 Tax=Hydra sp. RepID=Q00484_9CNID 62 4e-08
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 62 4e-08
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 62 4e-08
UniRef100_B7PJF9 Menin, putative n=1 Tax=Ixodes scapularis RepID... 62 4e-08
UniRef100_B7PAV3 Putative uncharacterized protein (Fragment) n=1... 62 4e-08
UniRef100_B4QT65 GD21099 n=1 Tax=Drosophila simulans RepID=B4QT6... 62 4e-08
UniRef100_B4MN04 GK17614 n=1 Tax=Drosophila willistoni RepID=B4M... 62 4e-08
UniRef100_B3P3J0 GG21917 n=1 Tax=Drosophila erecta RepID=B3P3J0_... 62 4e-08
UniRef100_B3MC79 GF12825 n=1 Tax=Drosophila ananassae RepID=B3MC... 62 4e-08
UniRef100_B3M2B3 GF17078 n=1 Tax=Drosophila ananassae RepID=B3M2... 62 4e-08
UniRef100_A8QF93 EBNA-2 nuclear protein, putative n=1 Tax=Brugia... 62 4e-08
UniRef100_A0NBM3 AGAP006897-PA n=1 Tax=Anopheles gambiae RepID=A... 62 4e-08
UniRef100_A5DSH8 Putative uncharacterized protein n=1 Tax=Lodder... 62 4e-08
UniRef100_Q86UP3 Zinc finger homeobox protein 4 n=1 Tax=Homo sap... 62 4e-08
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 61 5e-08
UniRef100_UPI0000D9F56F PREDICTED: similar to Protein CXorf45 n=... 61 5e-08
UniRef100_UPI000004873B protein kinase family protein n=1 Tax=Ar... 61 5e-08
UniRef100_UPI0001B7AE51 UPI0001B7AE51 related cluster n=1 Tax=Ra... 61 5e-08
UniRef100_UPI0001B7A7A5 UPI0001B7A7A5 related cluster n=1 Tax=Ra... 61 5e-08
UniRef100_A3F823 Mitochondrial fission regulator 1 n=1 Tax=Rattu... 61 5e-08
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 61 5e-08
UniRef100_A8GSX3 Putative uncharacterized protein n=1 Tax=Ricket... 61 5e-08
UniRef100_C5SP32 TonB protein n=1 Tax=Asticcacaulis excentricus ... 61 5e-08
UniRef100_Q9LS95 Somatic embryogenesis receptor kinase-like prot... 61 5e-08
UniRef100_O81106 Leucine-rich repeat transmembrane protein kinas... 61 5e-08
UniRef100_C5YU09 Putative uncharacterized protein Sb08g008380 n=... 61 5e-08
UniRef100_C4J566 Putative uncharacterized protein n=1 Tax=Zea ma... 61 5e-08
UniRef100_B9GW18 Predicted protein n=1 Tax=Populus trichocarpa R... 61 5e-08
UniRef100_Q5CKJ5 Putative uncharacterized protein n=1 Tax=Crypto... 61 5e-08
UniRef100_B7Q7F1 Secreted protein, putative (Fragment) n=1 Tax=I... 61 5e-08
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 61 5e-08
UniRef100_B4ND74 GK24962 n=1 Tax=Drosophila willistoni RepID=B4N... 61 5e-08
UniRef100_B4M7B5 GJ16487 n=1 Tax=Drosophila virilis RepID=B4M7B5... 61 5e-08
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 61 5e-08
UniRef100_B3N1Q5 GF20750 n=1 Tax=Drosophila ananassae RepID=B3N1... 61 5e-08
UniRef100_B2KTD4 Minicollagen 1 n=1 Tax=Clytia hemisphaerica Rep... 61 5e-08
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 61 5e-08
UniRef100_P0CB49 YLP motif-containing protein 1 n=1 Tax=Rattus n... 61 5e-08
UniRef100_Q9Z0G8-2 Isoform 2 of WAS/WASL-interacting protein fam... 61 5e-08
UniRef100_Q9Z0G8 WAS/WASL-interacting protein family member 3 n=... 61 5e-08
UniRef100_Q9AKJ0 Arp2/3 complex-activating protein rickA n=1 Tax... 61 5e-08
UniRef100_UPI0001B58635 hypothetical protein StAA4_25414 n=1 Tax... 61 7e-08
UniRef100_UPI000194BDE1 PREDICTED: zinc finger homeodomain 4 n=1... 61 7e-08
UniRef100_UPI0001926FBC PREDICTED: hypothetical protein n=1 Tax=... 61 7e-08
UniRef100_UPI0000F2C563 PREDICTED: similar to functional smad su... 61 7e-08
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 61 7e-08
UniRef100_UPI0000D9B690 PREDICTED: similar to diaphanous 1 n=1 T... 61 7e-08
UniRef100_UPI0001B7A6BD enabled homolog n=1 Tax=Rattus norvegicu... 61 7e-08
UniRef100_UPI0001B7A6BC enabled homolog n=1 Tax=Rattus norvegicu... 61 7e-08
UniRef100_UPI0001B7A6A7 enabled homolog n=1 Tax=Rattus norvegicu... 61 7e-08
UniRef100_UPI00015DEB7E YLP motif containing 1 n=1 Tax=Mus muscu... 61 7e-08
UniRef100_UPI00005A4477 PREDICTED: similar to zinc finger protei... 61 7e-08
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 61 7e-08
UniRef100_UPI0000ECD10B Zinc finger homeobox protein 4 (Zinc fin... 61 7e-08
UniRef100_Q4TB69 Chromosome 11 SCAF7190, whole genome shotgun se... 61 7e-08
UniRef100_Q4S280 Chromosome undetermined SCAF14764, whole genome... 61 7e-08
UniRef100_B4RDQ2 OmpA family protein n=1 Tax=Phenylobacterium zu... 61 7e-08
UniRef100_A5P9Z0 Peptidoglycan-associated protein n=1 Tax=Erythr... 61 7e-08
UniRef100_A3WEW6 Peptidoglycan-associated protein n=1 Tax=Erythr... 61 7e-08
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 61 7e-08
UniRef100_C1MU93 Predicted protein n=1 Tax=Micromonas pusilla CC... 61 7e-08
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 61 7e-08
UniRef100_A8MQH3 Uncharacterized protein At1g53730.2 n=1 Tax=Ara... 61 7e-08
UniRef100_A2XDP2 Putative uncharacterized protein n=1 Tax=Oryza ... 61 7e-08
UniRef100_B7Z9A8 cDNA FLJ58392 n=1 Tax=Homo sapiens RepID=B7Z9A8... 61 7e-08
UniRef100_Q4WDJ2 Cytokinesis protein SepA/Bni1 n=1 Tax=Aspergill... 61 7e-08
UniRef100_C8VTC6 Proline-rich, actin-associated protein Vrp1, pu... 61 7e-08
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 61 7e-08
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 61 7e-08
UniRef100_A5DK28 Putative uncharacterized protein n=1 Tax=Pichia... 61 7e-08
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 61 7e-08
UniRef100_Q9R0I7 YLP motif-containing protein 1 n=1 Tax=Mus musc... 61 7e-08
UniRef100_Q9C8M9 Protein STRUBBELIG-RECEPTOR FAMILY 6 n=2 Tax=Ar... 61 7e-08
UniRef100_Q10Q99 Formin-like protein 8 n=1 Tax=Oryza sativa Japo... 61 7e-08
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 60 9e-08
UniRef100_UPI000194C58F PREDICTED: formin 1 n=1 Tax=Taeniopygia ... 60 9e-08
UniRef100_UPI0001926BC8 PREDICTED: similar to mini-collagen isof... 60 9e-08
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 60 9e-08
UniRef100_UPI0000F2E99A PREDICTED: similar to diaphanous homolog... 60 9e-08
UniRef100_UPI000024DCC5 zinc finger homeodomain 4 n=1 Tax=Mus mu... 60 9e-08
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 60 9e-08
UniRef100_Q4RY48 Chromosome 3 SCAF14978, whole genome shotgun se... 60 9e-08
UniRef100_Q8PPF4 Putative uncharacterized protein n=1 Tax=Xantho... 60 9e-08
UniRef100_B4RES5 TonB protein n=1 Tax=Phenylobacterium zucineum ... 60 9e-08
UniRef100_B0BYE9 Arp2/3 complex activation protein n=1 Tax=Ricke... 60 9e-08
UniRef100_Q1NB62 OmpA/MotB n=1 Tax=Sphingomonas sp. SKA58 RepID=... 60 9e-08
UniRef100_Q9XIL9 Putative uncharacterized protein At2g15880 n=1 ... 60 9e-08
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 60 9e-08
UniRef100_Q079I0 Leucine-rich repeat transmembrane protein kinas... 60 9e-08
UniRef100_C6ZRX0 Leucine-rich repeat transmembrane protein kinas... 60 9e-08
UniRef100_C1PGW1 Tracheary element differentiation-related 7A n=... 60 9e-08
UniRef100_Q60EZ1 Os05g0207700 protein n=2 Tax=Oryza sativa RepID... 60 9e-08
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 60 9e-08
UniRef100_A7PVU3 Chromosome chr8 scaffold_34, whole genome shotg... 60 9e-08
UniRef100_A5BSD6 Putative uncharacterized protein n=1 Tax=Vitis ... 60 9e-08
UniRef100_Q00485 Mini-collagen (Fragment) n=1 Tax=Hydra sp. RepI... 60 9e-08
UniRef100_B4NSF9 GD15403 n=1 Tax=Drosophila simulans RepID=B4NSF... 60 9e-08
UniRef100_B4L5E2 GI21535 n=1 Tax=Drosophila mojavensis RepID=B4L... 60 9e-08
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 60 9e-08
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 60 9e-08
UniRef100_B2B5Z7 Predicted CDS Pa_2_6275 n=1 Tax=Podospora anser... 60 9e-08
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 60 9e-08
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI000151B9E8 hypothetical protein PGUG_05037 n=1 Tax=... 60 1e-07
UniRef100_UPI0000F202B4 PREDICTED: similar to tubby like protein... 60 1e-07
UniRef100_UPI0000519B2B PREDICTED: similar to CG2931-PA isoform ... 60 1e-07
UniRef100_UPI00001211EE Hypothetical protein CBG20761 n=1 Tax=Ca... 60 1e-07
UniRef100_UPI00004D5DA1 YLP motif containing protein 1 (Nuclear ... 60 1e-07
UniRef100_UPI00001809D7 UPI00001809D7 related cluster n=1 Tax=Ra... 60 1e-07
UniRef100_UPI00015DEA40 brain-specific angiogenesis inhibitor 1 ... 60 1e-07
UniRef100_UPI00016E72E1 UPI00016E72E1 related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI00016E5C2A UPI00016E5C2A related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_UPI0000ECB7C7 Formin-like protein 2 (Formin homology 2... 60 1e-07
UniRef100_Q4SUB2 Chromosome 3 SCAF13974, whole genome shotgun se... 60 1e-07
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 60 1e-07
UniRef100_Q6PB68 Bai1 protein (Fragment) n=1 Tax=Mus musculus Re... 60 1e-07
UniRef100_Q5DTN9 MKIAA4089 protein (Fragment) n=1 Tax=Mus muscul... 60 1e-07
UniRef100_B9EK88 Zinc finger protein 318 n=1 Tax=Mus musculus Re... 60 1e-07
UniRef100_B0V2M3 Zinc finger protein 318 n=3 Tax=Mus musculus Re... 60 1e-07
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 60 1e-07
UniRef100_Q0C0P5 OmpA family protein n=1 Tax=Hyphomonas neptuniu... 60 1e-07
UniRef100_B4W7V0 OmpA family protein n=1 Tax=Brevundimonas sp. B... 60 1e-07
UniRef100_A3WBR8 Peptidoglycan-associated protein n=1 Tax=Erythr... 60 1e-07
UniRef100_Q015R2 RhoA GTPase effector DIA/Diaphanous (ISS) n=1 T... 60 1e-07
UniRef100_C5XDG2 Putative uncharacterized protein Sb02g006780 n=... 60 1e-07
UniRef100_Q10SE3 Protein kinase domain containing protein, expre... 60 1e-07
UniRef100_Q4CLZ3 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_Q297D6 GA12408 n=1 Tax=Drosophila pseudoobscura pseudo... 60 1e-07
UniRef100_B9PQH8 Putative uncharacterized protein n=1 Tax=Toxopl... 60 1e-07
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 60 1e-07
UniRef100_B4KTK9 GI20587 n=1 Tax=Drosophila mojavensis RepID=B4K... 60 1e-07
UniRef100_B4IYS1 GH15108 n=1 Tax=Drosophila grimshawi RepID=B4IY... 60 1e-07
UniRef100_B4GE94 GL21863 n=1 Tax=Drosophila persimilis RepID=B4G... 60 1e-07
UniRef100_A8XYJ8 C. briggsae CBR-DDL-2 protein n=1 Tax=Caenorhab... 60 1e-07
UniRef100_A0E3T6 Chromosome undetermined scaffold_77, whole geno... 60 1e-07
UniRef100_Q5KHY4 Putative uncharacterized protein n=1 Tax=Filoba... 60 1e-07
UniRef100_Q0U2K7 Putative uncharacterized protein n=1 Tax=Phaeos... 60 1e-07
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 60 1e-07
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 60 1e-07
UniRef100_B9W9S7 Formin (Bud-site selection/polarity protein), p... 60 1e-07
UniRef100_Q99PP2 Zinc finger protein 318 (Fragment) n=1 Tax=Mus ... 60 1e-07
UniRef100_Q3UHD1 Brain-specific angiogenesis inhibitor 1 n=1 Tax... 60 1e-07
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 60 2e-07
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 60 2e-07
UniRef100_UPI000151AF18 hypothetical protein PGUG_03629 n=1 Tax=... 60 2e-07
UniRef100_UPI0000F2D288 PREDICTED: similar to eukaryotic transla... 60 2e-07
UniRef100_UPI0000E492FA PREDICTED: similar to L-delphilin n=1 Ta... 60 2e-07
UniRef100_UPI0000DD984C Os09g0538700 n=1 Tax=Oryza sativa Japoni... 60 2e-07
UniRef100_UPI00005A182D PREDICTED: similar to YLP motif containi... 60 2e-07
UniRef100_UPI00005A0B77 PREDICTED: similar to Vesicle-associated... 60 2e-07
UniRef100_UPI00005A063C PREDICTED: similar to junction-mediating... 60 2e-07
UniRef100_UPI000034EF33 SRF7 (STRUBBELIG-RECEPTOR FAMILY 7); ATP... 60 2e-07
UniRef100_UPI0001B7A397 jumonji domain containing 3 n=1 Tax=Ratt... 60 2e-07
UniRef100_UPI0000F4FC73 UPI0000F4FC73 related cluster n=1 Tax=Mu... 60 2e-07
UniRef100_UPI00016E597F UPI00016E597F related cluster n=1 Tax=Ta... 60 2e-07
UniRef100_UPI000195128A junction-mediating and regulatory protei... 60 2e-07
UniRef100_UPI0000ECBA12 Formin (Limb deformity protein). n=2 Tax... 60 2e-07
UniRef100_B0R0R1 Novel protein similar to human Ras association ... 60 2e-07
UniRef100_B1WBQ8 Glyceraldehyde 3-phosphate dehydrogenase n=1 Ta... 60 2e-07
UniRef100_A2A655 Bromodomain PHD finger transcription factor n=1... 60 2e-07
UniRef100_A2A654 Bromodomain PHD finger transcription factor n=1... 60 2e-07
UniRef100_Q3M5H7 VCBS n=1 Tax=Anabaena variabilis ATCC 29413 Rep... 60 2e-07
UniRef100_B0T768 TonB family protein n=1 Tax=Caulobacter sp. K31... 60 2e-07
UniRef100_A4TEZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 60 2e-07
UniRef100_A1W8T6 Nuclease (SNase domain protein) n=1 Tax=Acidovo... 60 2e-07
UniRef100_B4YB55 BimA n=1 Tax=Burkholderia pseudomallei RepID=B4... 60 2e-07
UniRef100_Q69JE5 Os09g0538700 protein n=1 Tax=Oryza sativa Japon... 60 2e-07
UniRef100_C1PGW2 Tracheary element differentiation-related 7B n=... 60 2e-07
UniRef100_B9IEK7 Predicted protein n=1 Tax=Populus trichocarpa R... 60 2e-07
UniRef100_B8BDX6 Putative uncharacterized protein n=1 Tax=Oryza ... 60 2e-07
UniRef100_A7XYI3 JXC1 n=1 Tax=Monodelphis domestica RepID=A7XYI3... 60 2e-07
UniRef100_Q7JW27 CG13176 n=1 Tax=Drosophila melanogaster RepID=Q... 60 2e-07
UniRef100_C4QFT9 Diaphanous, putative n=1 Tax=Schistosoma manson... 60 2e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 60 2e-07
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 60 2e-07
UniRef100_B4PVU3 GE23484 n=1 Tax=Drosophila yakuba RepID=B4PVU3_... 60 2e-07
UniRef100_B4NSF6 GD15401 n=1 Tax=Drosophila simulans RepID=B4NSF... 60 2e-07
UniRef100_B4HP59 GM21340 n=1 Tax=Drosophila sechellia RepID=B4HP... 60 2e-07
UniRef100_B3NTG0 GG18108 n=1 Tax=Drosophila erecta RepID=B3NTG0_... 60 2e-07
UniRef100_B3NS70 GG20253 n=1 Tax=Drosophila erecta RepID=B3NS70_... 60 2e-07
UniRef100_A2FMX2 DnaK protein n=1 Tax=Trichomonas vaginalis G3 R... 60 2e-07
UniRef100_A0BLV2 Chromosome undetermined scaffold_115, whole gen... 60 2e-07
UniRef100_C1H5C0 Decaprenyl-diphosphate synthase subunit 1 n=1 T... 60 2e-07
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 60 2e-07
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 60 2e-07
UniRef100_O93367 T-cell leukemia homeobox protein 3 n=1 Tax=Gall... 60 2e-07
UniRef100_Q9LUL4 Protein STRUBBELIG-RECEPTOR FAMILY 7 n=3 Tax=Ar... 60 2e-07
UniRef100_Q5NCY0 Lysine-specific demethylase 6B n=1 Tax=Mus musc... 60 2e-07
UniRef100_Q9ESV6 Glyceraldehyde-3-phosphate dehydrogenase, testi... 60 2e-07
UniRef100_Q54PI9 Formin-I n=1 Tax=Dictyostelium discoideum RepID... 60 2e-07
UniRef100_Q6ZQ03 Formin-binding protein 4 n=1 Tax=Mus musculus R... 60 2e-07
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 59 2e-07
UniRef100_UPI0001796908 PREDICTED: zinc finger homeobox 4 n=1 Ta... 59 2e-07
UniRef100_UPI0001552F36 PREDICTED: similar to SH3 domain binding... 59 2e-07
UniRef100_UPI0001552DAA PREDICTED: similar to CR16 n=1 Tax=Mus m... 59 2e-07
UniRef100_UPI0000E4921C PREDICTED: similar to formin 2 n=1 Tax=S... 59 2e-07
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 59 2e-07
UniRef100_UPI0000DA2E47 PREDICTED: similar to brain-specific ang... 59 2e-07
UniRef100_UPI0000DA2D13 PREDICTED: similar to brain-specific ang... 59 2e-07
UniRef100_UPI0000D9E82E PREDICTED: similar to additional sex com... 59 2e-07
UniRef100_UPI0000D9E82D PREDICTED: similar to additional sex com... 59 2e-07
UniRef100_UPI0000547F81 PREDICTED: formin 1 n=1 Tax=Danio rerio ... 59 2e-07
UniRef100_UPI00003BF9F1 PREDICTED: similar to SCAR CG4636-PA n=1... 59 2e-07
UniRef100_UPI0001B7AE1B UPI0001B7AE1B related cluster n=1 Tax=Ra... 59 2e-07
UniRef100_UPI0000503F16 UPI0000503F16 related cluster n=1 Tax=Ra... 59 2e-07