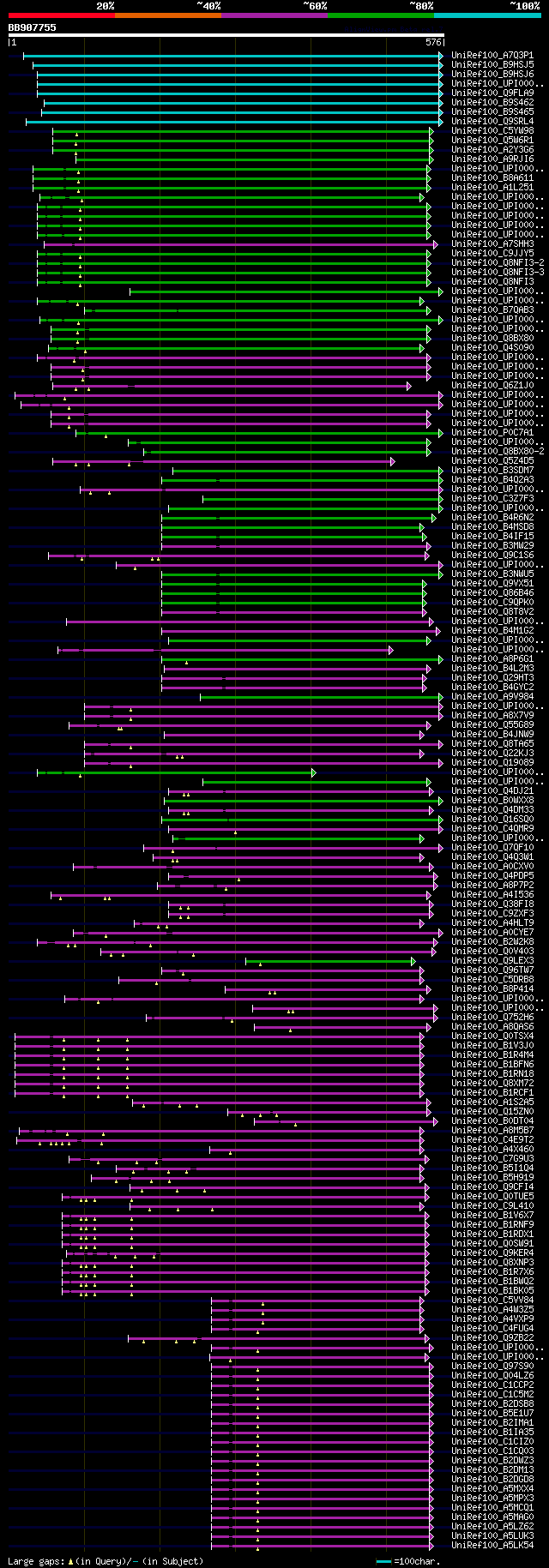
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB907755 RCE06152
(576 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_A7Q3P1 Chromosome chr13 scaffold_48, whole genome shot... 310 5e-83
UniRef100_B9HSJ5 Predicted protein n=1 Tax=Populus trichocarpa R... 306 6e-82
UniRef100_B9HSJ6 Predicted protein n=1 Tax=Populus trichocarpa R... 304 3e-81
UniRef100_UPI00005DC273 hydrolase, acting on glycosyl bonds / ma... 286 1e-75
UniRef100_Q9FLA9 Similarity to endo-beta-N-acetylglucosaminidase... 286 1e-75
UniRef100_B9S462 Endo beta n-acetylglucosaminidase, putative n=1... 286 1e-75
UniRef100_B9S465 Endo beta n-acetylglucosaminidase, putative n=1... 285 2e-75
UniRef100_Q9SRL4 Putative uncharacterized protein F9F8.14 n=1 Ta... 273 5e-72
UniRef100_C5YW98 Putative uncharacterized protein Sb09g016230 n=... 247 5e-64
UniRef100_Q5W6R1 Os05g0346500 protein n=3 Tax=Oryza sativa Japon... 236 9e-61
UniRef100_A2Y3G6 Putative uncharacterized protein n=1 Tax=Oryza ... 236 9e-61
UniRef100_A9RJI6 Predicted protein n=1 Tax=Physcomitrella patens... 191 3e-47
UniRef100_UPI0001A2D628 hypothetical protein LOC561239 n=1 Tax=D... 160 8e-38
UniRef100_B8A611 Novel protein with a Glycosyl hydrolase family ... 160 8e-38
UniRef100_A1L251 Cytosolic endo-beta-N-acetylglucosaminidase n=1... 159 1e-37
UniRef100_UPI00016E76D3 UPI00016E76D3 related cluster n=1 Tax=Ta... 155 2e-36
UniRef100_UPI0000E24B20 PREDICTED: hypothetical protein isoform ... 153 1e-35
UniRef100_UPI0000E24B1F PREDICTED: endo-beta-N-acetylglucosamini... 153 1e-35
UniRef100_UPI0000E24B1E PREDICTED: endo-beta-N-acetylglucosamini... 153 1e-35
UniRef100_UPI0000D9E532 PREDICTED: similar to endo-beta-N-acetyl... 153 1e-35
UniRef100_A7SHH3 Predicted protein (Fragment) n=1 Tax=Nematostel... 153 1e-35
UniRef100_C9JJY5 Putative uncharacterized protein ENGASE n=1 Tax... 153 1e-35
UniRef100_Q8NFI3-2 Isoform 2 of Cytosolic endo-beta-N-acetylgluc... 153 1e-35
UniRef100_Q8NFI3-3 Isoform 3 of Cytosolic endo-beta-N-acetylgluc... 153 1e-35
UniRef100_Q8NFI3 Cytosolic endo-beta-N-acetylglucosaminidase n=1... 153 1e-35
UniRef100_UPI0001866D1E hypothetical protein BRAFLDRAFT_227310 n... 152 1e-35
UniRef100_UPI00017B4728 UPI00017B4728 related cluster n=1 Tax=Te... 152 2e-35
UniRef100_B7QAB3 Endo beta N-acetylglucosaminidase, putative (Fr... 152 2e-35
UniRef100_UPI0000F2C04D PREDICTED: similar to endo-beta-N-acetyl... 151 3e-35
UniRef100_UPI00001D068C endo-beta-N-acetylglucosaminidase n=1 Ta... 151 3e-35
UniRef100_Q8BX80 Cytosolic endo-beta-N-acetylglucosaminidase n=1... 150 9e-35
UniRef100_Q4S090 Chromosome undetermined SCAF14784, whole genome... 149 2e-34
UniRef100_UPI0001796AD0 PREDICTED: similar to endo-beta-N-acetyl... 148 3e-34
UniRef100_UPI00005A1A59 PREDICTED: similar to RIKEN cDNA D230014... 147 6e-34
UniRef100_UPI0000EB1EFC endo-beta-N-acetylglucosaminidase n=1 Ta... 147 6e-34
UniRef100_Q6Z1J0 Glycosyl hydrolase family 85-like protein n=1 T... 146 1e-33
UniRef100_UPI0000E812E5 PREDICTED: similar to endo-beta-N-acetyl... 145 2e-33
UniRef100_UPI0000ECA5AA endo-beta-N-acetylglucosaminidase n=1 Ta... 144 4e-33
UniRef100_UPI00017C385F PREDICTED: similar to endo-beta-N-acetyl... 144 5e-33
UniRef100_UPI0000EBDE7A UPI0000EBDE7A related cluster n=1 Tax=Bo... 144 5e-33
UniRef100_P0C7A1 Cytosolic endo-beta-N-acetylglucosaminidase n=1... 144 5e-33
UniRef100_UPI0001B7A419 UPI0001B7A419 related cluster n=1 Tax=Ra... 143 1e-32
UniRef100_Q8BX80-2 Isoform 2 of Cytosolic endo-beta-N-acetylgluc... 142 1e-32
UniRef100_Q5Z4D5 Glycosyl hydrolase family 85-like protein n=1 T... 141 3e-32
UniRef100_B3SDM7 Putative uncharacterized protein (Fragment) n=1... 135 2e-30
UniRef100_B4Q2A3 GE17667 n=1 Tax=Drosophila yakuba RepID=B4Q2A3_... 132 2e-29
UniRef100_UPI0000DB751B PREDICTED: similar to CG5613-PA, isoform... 130 9e-29
UniRef100_C3Z7F3 Putative uncharacterized protein (Fragment) n=1... 129 2e-28
UniRef100_UPI00015B5070 PREDICTED: hypothetical protein n=1 Tax=... 128 3e-28
UniRef100_B4R6N2 GD15670 n=1 Tax=Drosophila simulans RepID=B4R6N... 128 4e-28
UniRef100_B4MSD8 GK19954 n=1 Tax=Drosophila willistoni RepID=B4M... 127 6e-28
UniRef100_B4IF15 GM13512 n=1 Tax=Drosophila sechellia RepID=B4IF... 127 6e-28
UniRef100_B3MW29 GF22338 n=1 Tax=Drosophila ananassae RepID=B3MW... 127 8e-28
UniRef100_Q9C1S6 Endo-b-N-acetylglucosaminidase n=1 Tax=Mucor hi... 126 1e-27
UniRef100_UPI0001791C18 PREDICTED: similar to endo-beta-N-acetyl... 126 1e-27
UniRef100_B3NWU5 GG19117 n=1 Tax=Drosophila erecta RepID=B3NWU5_... 126 1e-27
UniRef100_Q9VX51 CG5613, isoform A n=1 Tax=Drosophila melanogast... 125 3e-27
UniRef100_Q86B46 CG5613, isoform B n=1 Tax=Drosophila melanogast... 125 3e-27
UniRef100_C9QPK0 RH12325p (Fragment) n=1 Tax=Drosophila melanoga... 125 3e-27
UniRef100_Q8T8V2 AT22312p n=1 Tax=Drosophila melanogaster RepID=... 124 4e-27
UniRef100_UPI000180C583 PREDICTED: similar to predicted protein ... 124 7e-27
UniRef100_B4M1G2 GJ18856 n=1 Tax=Drosophila virilis RepID=B4M1G2... 124 7e-27
UniRef100_UPI0000D56E54 PREDICTED: similar to RIKEN cDNA D230014... 123 9e-27
UniRef100_UPI0000E48D8F PREDICTED: similar to endo-beta-N-acetyl... 123 1e-26
UniRef100_A8P6G1 Glycosyl hydrolase family 85 protein n=1 Tax=Br... 123 1e-26
UniRef100_B4L2M3 GI15965 n=1 Tax=Drosophila mojavensis RepID=B4L... 122 2e-26
UniRef100_Q29HT3 GA19007 n=1 Tax=Drosophila pseudoobscura pseudo... 121 3e-26
UniRef100_B4GYC2 GL19891 n=1 Tax=Drosophila persimilis RepID=B4G... 121 3e-26
UniRef100_A9V984 Predicted protein n=1 Tax=Monosiga brevicollis ... 119 2e-25
UniRef100_UPI0000220A6F hypothetical protein CBG09066 n=1 Tax=Ca... 119 2e-25
UniRef100_A8X7V9 C. briggsae CBR-ENG-1 protein n=1 Tax=Caenorhab... 119 2e-25
UniRef100_Q55G89 Putative uncharacterized protein n=1 Tax=Dictyo... 118 4e-25
UniRef100_B4JNW9 GH24904 n=1 Tax=Drosophila grimshawi RepID=B4JN... 116 1e-24
UniRef100_Q8TA65 Endo-beta-N-acetylglucosaminidase n=2 Tax=Caeno... 115 2e-24
UniRef100_Q22KJ3 Glycosyl hydrolase family 85 protein n=1 Tax=Te... 115 2e-24
UniRef100_Q19089 Putative uncharacterized protein n=1 Tax=Caenor... 115 2e-24
UniRef100_UPI0001556197 PREDICTED: similar to endo-beta-N-acetyl... 111 5e-23
UniRef100_UPI00019256CF PREDICTED: similar to endo-beta-N-acetyl... 110 6e-23
UniRef100_Q4DJ21 Endo-beta-N-acetylglucosaminidase, putative n=1... 109 2e-22
UniRef100_B0WXX8 Endo beta N-acetyl glucosaminidase n=1 Tax=Cule... 107 5e-22
UniRef100_Q4DM33 Endo-beta-N-acetylglucosaminidase, putative n=1... 107 7e-22
UniRef100_Q16SQ0 Endo beta n-acetylglucosaminidase n=1 Tax=Aedes... 105 2e-21
UniRef100_C4QMR9 Endo beta n-acetylglucosaminidase, putative n=1... 104 6e-21
UniRef100_UPI000186D06C endo beta N-acetylglucosaminidase, putat... 103 7e-21
UniRef100_Q7QF10 AGAP000249-PA (Fragment) n=1 Tax=Anopheles gamb... 102 2e-20
UniRef100_Q4Q3W1 Glycosyl hydrolase-like protein n=1 Tax=Leishma... 100 8e-20
UniRef100_A0CXV0 Chromosome undetermined scaffold_30, whole geno... 99 2e-19
UniRef100_Q4PDP5 Putative uncharacterized protein n=1 Tax=Ustila... 99 3e-19
UniRef100_A8P7P2 Putative uncharacterized protein n=1 Tax=Coprin... 99 3e-19
UniRef100_A4I536 Glycosyl hydrolase-like protein n=1 Tax=Leishma... 97 1e-18
UniRef100_Q38FI8 Endo-beta-N-acetylglucosaminidase, putative n=1... 96 2e-18
UniRef100_C9ZXF3 Endo-beta-N-acetylglucosaminidase, putative n=1... 96 2e-18
UniRef100_A4HLT9 Glycosyl hydrolase-like protein n=1 Tax=Leishma... 96 2e-18
UniRef100_A0CYE7 Chromosome undetermined scaffold_31, whole geno... 96 2e-18
UniRef100_B2W2K8 Glycosyl hydrolase family 85 protein n=1 Tax=Py... 93 1e-17
UniRef100_Q0V403 Putative uncharacterized protein n=1 Tax=Phaeos... 93 2e-17
UniRef100_Q9LEX3 Putative uncharacterized protein T27I15_100 n=1... 92 2e-17
UniRef100_Q96TW7 Putative uncharacterized protein n=1 Tax=Wicker... 90 1e-16
UniRef100_C5DRB8 ZYRO0B07216p n=1 Tax=Zygosaccharomyces rouxii C... 89 2e-16
UniRef100_B8P414 Putative uncharacterized protein n=1 Tax=Postia... 79 3e-13
UniRef100_UPI00004D2529 endo-beta-N-acetylglucosaminidase n=1 Ta... 77 7e-13
UniRef100_UPI000187D79C hypothetical protein MPER_11065 n=1 Tax=... 76 2e-12
UniRef100_Q752H6 AFR597Wp n=1 Tax=Eremothecium gossypii RepID=Q7... 76 2e-12
UniRef100_A8QAS6 Putative uncharacterized protein n=1 Tax=Malass... 72 4e-11
UniRef100_Q0TSX4 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 70 9e-11
UniRef100_B1V3J0 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 70 9e-11
UniRef100_B1R4M4 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 70 9e-11
UniRef100_B1BFN6 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 70 9e-11
UniRef100_B1RN18 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 70 1e-10
UniRef100_Q8XM72 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 68 4e-10
UniRef100_B1RCF1 Glycosyl hydrolase, family 85 n=1 Tax=Clostridi... 68 4e-10
UniRef100_A1S2A5 Glycoside hydrolase, family 85 n=1 Tax=Shewanel... 65 3e-09
UniRef100_Q15ZN0 Glycoside hydrolase, family 85 n=1 Tax=Pseudoal... 65 4e-09
UniRef100_B0DT04 Glycoside hydrolase family 85 protein n=1 Tax=L... 65 5e-09
UniRef100_A8M5B7 Glycoside hydrolase family 85 n=1 Tax=Salinispo... 64 8e-09
UniRef100_C4E9T2 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 64 1e-08
UniRef100_A4X460 Glycoside hydrolase, family 85 n=1 Tax=Salinisp... 63 1e-08
UniRef100_C7G9U3 Mannosyl-glycoprotein endo-beta-N-acetylglucosa... 63 2e-08
UniRef100_B5I1Q4 Endo-beta-N-acetylglucosaminidase n=1 Tax=Strep... 62 3e-08
UniRef100_B5H919 Endo-beta-N-acetylglucosaminidase n=1 Tax=Strep... 60 2e-07
UniRef100_Q9CFI4 Endo-beta-N-acetylglucosaminidase n=1 Tax=Lacto... 59 2e-07
UniRef100_Q0TUE5 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 57 8e-07
UniRef100_C9L410 Glycosyl hydrolase, family 85 n=1 Tax=Blautia h... 57 8e-07
UniRef100_B1V6X7 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 57 8e-07
UniRef100_B1RNF9 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 57 8e-07
UniRef100_B1RDX1 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 57 8e-07
UniRef100_Q0SW91 Glycosyl hydrolase family 85 family n=1 Tax=Clo... 57 1e-06
UniRef100_Q9KER4 Endo-beta-N-acetylglucosaminidase n=1 Tax=Bacil... 56 2e-06
UniRef100_Q8XNP3 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 56 2e-06
UniRef100_B1R7X6 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 56 2e-06
UniRef100_B1BWQ2 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 56 2e-06
UniRef100_B1BK05 Endo-beta-N-acetylglucosaminidase n=1 Tax=Clost... 56 2e-06
UniRef100_C5VV84 Putative endo-beta-N-acetylglucosaminidase n=1 ... 56 2e-06
UniRef100_A4W3Z5 Endo-beta-N-acetylglucosaminidase, putative n=1... 56 2e-06
UniRef100_A4VXP9 Endo-beta-N-acetylglucosaminidase, putative n=3... 56 2e-06
UniRef100_C4FUG4 Putative uncharacterized protein n=1 Tax=Catone... 55 3e-06
UniRef100_Q9ZB22 Endo-beta-N-acetylglucosaminidase n=1 Tax=Arthr... 55 5e-06
UniRef100_UPI0001AF4DB7 endo-beta-N-acetylglucosaminidase, putat... 54 9e-06
UniRef100_UPI0001789033 Mannosyl-glycoprotein endo-beta-N-acetyl... 54 9e-06
UniRef100_Q97S90 Putative endo-beta-N-acetylglucosaminidase n=1 ... 54 9e-06
UniRef100_Q04LZ6 Endo-beta-N-acetylglucosaminidase, putative n=2... 54 9e-06
UniRef100_C1CCP2 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_C1C5M2 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_B2DSB8 Endo-beta-N-acetylglucosaminidase D n=2 Tax=Str... 54 9e-06
UniRef100_B5E1U7 Endo-beta-N-acetylglucosaminidase, point mutati... 54 9e-06
UniRef100_B2IMA1 Endo-beta-N-acetylglucosaminidase, putative n=1... 54 9e-06
UniRef100_B1IA35 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_C1CIZ0 Endo-beta-N-acetylglucosaminidase D n=2 Tax=Str... 54 9e-06
UniRef100_C1CQ03 Endo-beta-N-acetylglucosaminidase D n=2 Tax=Str... 54 9e-06
UniRef100_B2DWZ3 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_B2DM13 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_B2DGD8 Endo-beta-N-acetylglucosaminidase D n=1 Tax=Str... 54 9e-06
UniRef100_A5MXX4 CTP synthetase n=1 Tax=Streptococcus pneumoniae... 54 9e-06
UniRef100_A5MPX3 Endo-beta-N-acetylglucosaminidase, putative n=1... 54 9e-06
UniRef100_A5MCQ1 CTP synthetase n=1 Tax=Streptococcus pneumoniae... 54 9e-06
UniRef100_A5MAG0 CTP synthetase n=1 Tax=Streptococcus pneumoniae... 54 9e-06
UniRef100_A5LZ62 CTP synthetase n=2 Tax=Streptococcus pneumoniae... 54 9e-06
UniRef100_A5LUK3 Endo-beta-N-acetylglucosaminidase, putative n=1... 54 9e-06
UniRef100_A5LK54 CTP synthetase n=1 Tax=Streptococcus pneumoniae... 54 9e-06
UniRef100_A5LA50 CTP synthetase n=1 Tax=Streptococcus pneumoniae... 54 9e-06