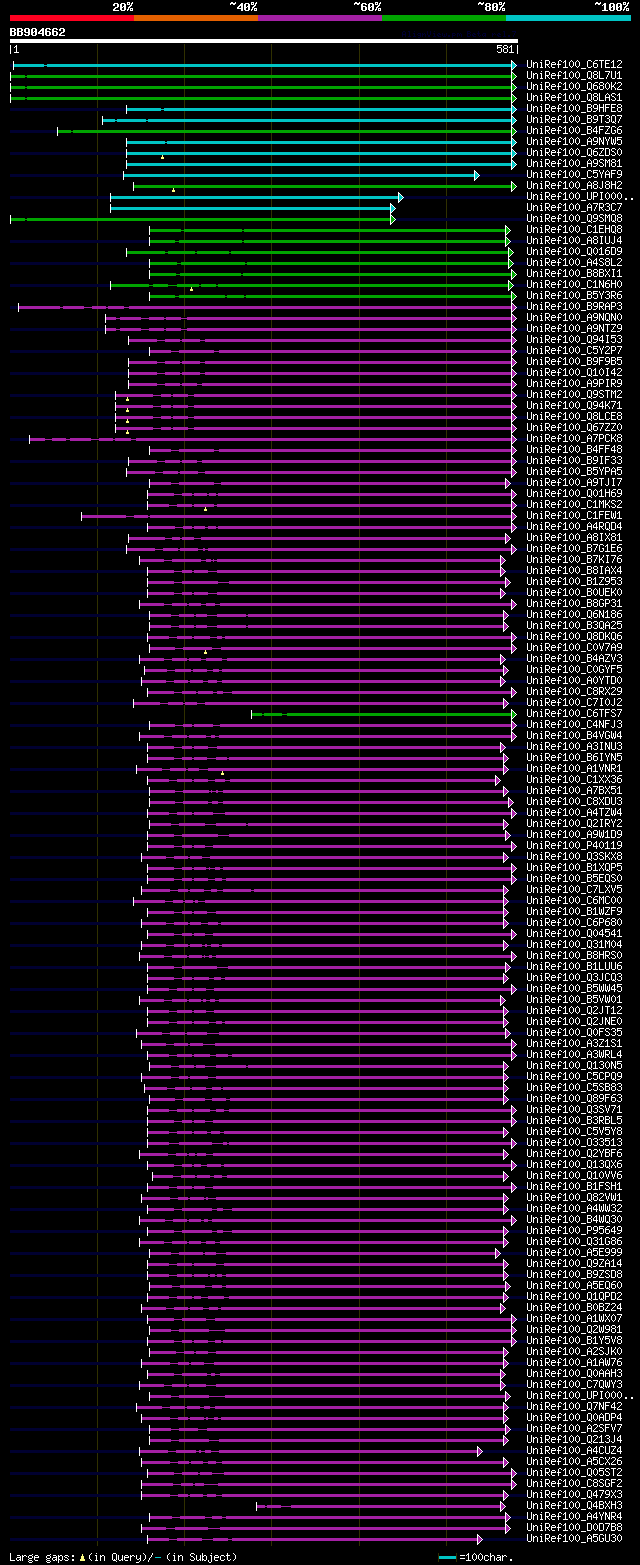
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BB904662 RCE02630
(581 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_C6TE12 Putative uncharacterized protein n=1 Tax=Glycin... 300 4e-80
UniRef100_Q8L7U1 AT4g39970/T5J17_140 n=1 Tax=Arabidopsis thalian... 257 5e-67
UniRef100_Q680K2 mRNA, clone: RAFL22-48-I16 n=1 Tax=Arabidopsis ... 257 5e-67
UniRef100_Q8LAS1 Putative uncharacterized protein n=1 Tax=Arabid... 255 2e-66
UniRef100_B9HFE8 Predicted protein (Fragment) n=1 Tax=Populus tr... 251 4e-65
UniRef100_B9T3Q7 2-deoxyglucose-6-phosphate phosphatase, putativ... 249 1e-64
UniRef100_B4FZG6 Putative uncharacterized protein n=1 Tax=Zea ma... 234 5e-60
UniRef100_A9NYW5 Putative uncharacterized protein n=1 Tax=Picea ... 231 2e-59
UniRef100_Q6ZDS0 Os08g0485900 protein n=2 Tax=Oryza sativa RepID... 229 9e-59
UniRef100_A9SM81 Predicted protein n=1 Tax=Physcomitrella patens... 218 3e-55
UniRef100_C5YAF9 Putative uncharacterized protein Sb06g032650 n=... 216 1e-54
UniRef100_A8J8H2 Predicted protein n=1 Tax=Chlamydomonas reinhar... 192 2e-47
UniRef100_UPI0001985F77 PREDICTED: hypothetical protein n=1 Tax=... 177 5e-43
UniRef100_A7R3C7 Chromosome undetermined scaffold_502, whole gen... 176 2e-42
UniRef100_Q9SMQ8 Putative uncharacterized protein AT4g39970 n=1 ... 174 3e-42
UniRef100_C1EHQ8 Predicted protein (Fragment) n=1 Tax=Micromonas... 146 1e-33
UniRef100_A8IUJ4 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 144 4e-33
UniRef100_Q016D9 Predicted haloacid-halidohydrolase and related ... 144 6e-33
UniRef100_A4S8L2 Predicted protein n=1 Tax=Ostreococcus lucimari... 143 1e-32
UniRef100_B8BXI1 Predicted protein (Fragment) n=1 Tax=Thalassios... 119 2e-25
UniRef100_C1N6H0 Predicted protein n=1 Tax=Micromonas pusilla CC... 118 4e-25
UniRef100_B5Y3R6 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 114 7e-24
UniRef100_B9RAP3 2-deoxyglucose-6-phosphate phosphatase, putativ... 98 4e-19
UniRef100_A9NQN0 Putative uncharacterized protein n=1 Tax=Picea ... 97 7e-19
UniRef100_A9NTZ9 Putative uncharacterized protein n=1 Tax=Picea ... 97 1e-18
UniRef100_Q94I53 Putative hydrolase n=1 Tax=Oryza sativa Japonic... 96 2e-18
UniRef100_C5Y2P7 Putative uncharacterized protein Sb05g018080 n=... 96 2e-18
UniRef100_B9F9B5 Putative uncharacterized protein n=1 Tax=Oryza ... 96 2e-18
UniRef100_Q10I42 Os03g0565200 protein n=2 Tax=Oryza sativa RepID... 96 2e-18
UniRef100_A9PIR9 Putative uncharacterized protein n=1 Tax=Populu... 94 6e-18
UniRef100_Q9STM2 Putative uncharacterized protein T29H11_60 n=1 ... 94 1e-17
UniRef100_Q94K71 Putative uncharacterized protein At3g48420 n=1 ... 94 1e-17
UniRef100_Q8LCE8 Putative uncharacterized protein n=1 Tax=Arabid... 94 1e-17
UniRef100_Q67ZZ0 Putative uncharacterized protein At3g48415 n=1 ... 94 1e-17
UniRef100_A7PCK8 Chromosome chr17 scaffold_12, whole genome shot... 94 1e-17
UniRef100_B4FF48 Protein cbbY n=1 Tax=Zea mays RepID=B4FF48_MAIZE 93 2e-17
UniRef100_B9IF33 Predicted protein n=1 Tax=Populus trichocarpa R... 92 4e-17
UniRef100_B5YPA5 Possibly a phosphatase from the CbbY protein fa... 91 7e-17
UniRef100_A9TJI7 Predicted protein n=1 Tax=Physcomitrella patens... 91 8e-17
UniRef100_Q01H69 Predicted haloacid-halidohydrolase and related ... 90 1e-16
UniRef100_C1MKS2 Predicted protein n=1 Tax=Micromonas pusilla CC... 90 1e-16
UniRef100_C1FEW1 Haloacid-halidohydrolase n=1 Tax=Micromonas sp.... 87 7e-16
UniRef100_A4RQD4 Predicted protein (Fragment) n=1 Tax=Ostreococc... 86 3e-15
UniRef100_A8IX81 Predicted protein n=1 Tax=Chlamydomonas reinhar... 80 1e-13
UniRef100_B7G1E6 Predicted protein (Fragment) n=1 Tax=Phaeodacty... 79 2e-13
UniRef100_B7KI76 HAD-superfamily hydrolase, subfamily IA, varian... 77 7e-13
UniRef100_B8IAX4 HAD-superfamily hydrolase, subfamily IA, varian... 77 1e-12
UniRef100_B1Z953 HAD-superfamily hydrolase, subfamily IA, varian... 75 3e-12
UniRef100_B0UEK0 HAD-superfamily hydrolase, subfamily IA, varian... 75 3e-12
UniRef100_B8GP31 HAD-superfamily hydrolase, subfamily IA, varian... 75 4e-12
UniRef100_Q6N186 Haloacid dehalogenase-like hydrolase n=1 Tax=Rh... 74 6e-12
UniRef100_B3QA25 HAD-superfamily hydrolase, subfamily IA, varian... 74 6e-12
UniRef100_Q8DKQ6 CbbY family protein n=1 Tax=Thermosynechococcus... 74 8e-12
UniRef100_C0V7A9 Haloacid dehalogenase superfamily protein, subf... 74 1e-11
UniRef100_B4AZV3 HAD-superfamily hydrolase, subfamily IA, varian... 73 1e-11
UniRef100_C0GYF5 HAD-superfamily hydrolase, subfamily IA, varian... 73 2e-11
UniRef100_A0YTD0 CbbY family protein n=1 Tax=Lyngbya sp. PCC 810... 73 2e-11
UniRef100_C8RX29 HAD-superfamily hydrolase, subfamily IA, varian... 72 2e-11
UniRef100_C7I0J2 HAD-superfamily hydrolase, subfamily IA, varian... 72 2e-11
UniRef100_C6TFS7 Putative uncharacterized protein n=1 Tax=Glycin... 72 2e-11
UniRef100_C4NFJ3 Putative hydrolase (Fragment) n=1 Tax=Actinopla... 72 4e-11
UniRef100_B4VGW4 Haloacid dehalogenase-like hydrolase, putative ... 72 4e-11
UniRef100_A3INU3 HAD-superfamily hydrolase, subfamily IA, varian... 72 4e-11
UniRef100_B6IYN5 CbbY protein n=1 Tax=Rhodospirillum centenum SW... 71 5e-11
UniRef100_A1VNR1 HAD-superfamily hydrolase, subfamily IA, varian... 71 5e-11
UniRef100_C1XX36 Haloacid dehalogenase superfamily protein, subf... 71 5e-11
UniRef100_A7BX51 HAD-superfamily hydrolase, subfamily IA, varian... 71 5e-11
UniRef100_C8XDU3 HAD-superfamily hydrolase, subfamily IA, varian... 71 7e-11
UniRef100_A4TZW4 HAD-superfamily hydrolase subfamily IA, variant... 71 7e-11
UniRef100_Q2IRY2 HAD-superfamily hydrolase subfamily IA, variant... 70 9e-11
UniRef100_A9W1D9 HAD-superfamily hydrolase, subfamily IA, varian... 70 9e-11
UniRef100_P40119 Protein cbbY, chromosomal n=1 Tax=Ralstonia eut... 70 1e-10
UniRef100_Q3SKX8 HAD-superfamily hydrolase subfamily IA, variant... 70 2e-10
UniRef100_B1XQP5 CbbY family protein n=1 Tax=Synechococcus sp. P... 69 3e-10
UniRef100_B5EQS0 HAD-superfamily hydrolase, subfamily IA, varian... 69 3e-10
UniRef100_C7LXV5 HAD-superfamily hydrolase, subfamily IA, varian... 68 5e-10
UniRef100_C6MC00 HAD-superfamily hydrolase, subfamily IA, varian... 68 5e-10
UniRef100_B1WZF9 HAD-superfamily hydrolase, subfamily IA, varian... 68 6e-10
UniRef100_C6P680 HAD-superfamily hydrolase, subfamily IA, varian... 68 6e-10
UniRef100_Q04541 Protein cbbY, plasmid n=1 Tax=Ralstonia eutroph... 68 6e-10
UniRef100_Q31M04 HAD-superfamily hydrolase subfamily IA, variant... 67 8e-10
UniRef100_B8HRS0 HAD-superfamily hydrolase, subfamily IA, varian... 67 8e-10
UniRef100_B1LUU6 HAD-superfamily hydrolase, subfamily IA, varian... 67 8e-10
UniRef100_Q3JCQ3 HAD-superfamily hydrolase subfamily IA, variant... 67 8e-10
UniRef100_B5WW45 HAD-superfamily hydrolase, subfamily IA, varian... 67 8e-10
UniRef100_B5VW01 HAD-superfamily hydrolase, subfamily IA, varian... 67 8e-10
UniRef100_Q2JT12 HAD-superfamily hydrolase, subfamily IA, varian... 67 1e-09
UniRef100_Q2JNE0 HAD-superfamily hydrolase, subfamily IA, varian... 67 1e-09
UniRef100_Q0FS35 CbbY family protein n=1 Tax=Roseovarius sp. HTC... 66 2e-09
UniRef100_A3Z1S1 Probable hydrolase, CbbY protein-like n=1 Tax=S... 66 2e-09
UniRef100_A3WRL4 HAD-superfamily hydrolase subfamily IA, variant... 66 2e-09
UniRef100_Q130N5 HAD-superfamily hydrolase subfamily IA, variant... 66 2e-09
UniRef100_C5CPQ9 HAD-superfamily hydrolase, subfamily IA, varian... 66 2e-09
UniRef100_C5SB83 HAD-superfamily hydrolase, subfamily IA, varian... 66 2e-09
UniRef100_Q89F63 CbbY protein n=1 Tax=Bradyrhizobium japonicum R... 65 3e-09
UniRef100_Q3SV71 HAD-superfamily hydrolase subfamily IA, variant... 65 3e-09
UniRef100_B3RBL5 Putative uncharacterized protein cbbY n=1 Tax=C... 65 4e-09
UniRef100_C5V5Y8 HAD-superfamily hydrolase, subfamily IA, varian... 65 4e-09
UniRef100_O33513 Protein cbbY n=1 Tax=Rhodobacter capsulatus Rep... 65 4e-09
UniRef100_Q2YBF6 HAD-superfamily hydrolase subfamily IA, variant... 65 5e-09
UniRef100_Q13QX6 HAD-superfamily hydrolase, subfamily IA, varian... 65 5e-09
UniRef100_Q10VV6 HAD-superfamily hydrolase, subfamily IA, varian... 65 5e-09
UniRef100_B1FSH1 HAD-superfamily hydrolase, subfamily IA, varian... 65 5e-09
UniRef100_Q82VW1 Hydrolase family n=1 Tax=Nitrosomonas europaea ... 64 7e-09
UniRef100_A4WW32 HAD-superfamily hydrolase, subfamily IA, varian... 64 7e-09
UniRef100_B4WQ30 Haloacid dehalogenase-like hydrolase, putative ... 64 9e-09
UniRef100_P95649 Protein cbbY n=4 Tax=Rhodobacter sphaeroides Re... 64 9e-09
UniRef100_Q31G86 Haloacid dehalogenase-like hydrolase family pro... 64 1e-08
UniRef100_A5E999 Putative haloacid dehalogenase-like hydrolase c... 64 1e-08
UniRef100_Q9ZA14 CbbY n=1 Tax=Hydrogenophilus thermoluteolus Rep... 64 1e-08
UniRef100_B9ZSD8 HAD-superfamily hydrolase, subfamily IA, varian... 64 1e-08
UniRef100_A5EQ60 Putative haloacid dehalogenase-like hydrolase, ... 63 1e-08
UniRef100_Q1QPD2 HAD-superfamily hydrolase subfamily IA, variant... 63 2e-08
UniRef100_B0BZ24 HAD-superfamily hydrolase, subfamily IA, varian... 63 2e-08
UniRef100_A1WX07 HAD-superfamily hydrolase, subfamily IA, varian... 62 2e-08
UniRef100_Q2W981 CbbY protein n=1 Tax=Magnetospirillum magneticu... 62 3e-08
UniRef100_B1Y5V8 HAD-superfamily hydrolase, subfamily IA, varian... 62 3e-08
UniRef100_A2SJK0 Haloacid dehalogenase n=1 Tax=Methylibium petro... 62 3e-08
UniRef100_A1AW76 HAD-superfamily hydrolase, subfamily IA, varian... 62 3e-08
UniRef100_Q0AAH3 HAD-superfamily hydrolase, subfamily IA, varian... 62 4e-08
UniRef100_C7QWY3 HAD-superfamily hydrolase, subfamily IA, varian... 62 4e-08
UniRef100_UPI000038439E COG0637: Predicted phosphatase/phosphohe... 61 6e-08
UniRef100_Q7NF42 Glr3684 protein n=1 Tax=Gloeobacter violaceus R... 61 6e-08
UniRef100_Q0ADP4 Haloacid dehalogenase domain protein hydrolase ... 61 7e-08
UniRef100_A2SFV7 Haloacid dehalogenase n=1 Tax=Methylibium petro... 61 7e-08
UniRef100_Q213J4 HAD-superfamily hydrolase subfamily IA, variant... 60 1e-07
UniRef100_A4CUZ4 Putative uncharacterized protein n=1 Tax=Synech... 59 2e-07
UniRef100_A5CX26 HAD-superfamily hydrolase, subfamily IA n=1 Tax... 59 3e-07
UniRef100_Q05ST2 Putative uncharacterized protein n=1 Tax=Synech... 59 3e-07
UniRef100_C8SGF2 HAD-superfamily hydrolase, subfamily IA, varian... 59 4e-07
UniRef100_Q479X3 HAD-superfamily hydrolase subfamily IA, variant... 57 8e-07
UniRef100_Q4BXH3 HAD-superfamily hydrolase, subfamily IA, varian... 57 1e-06
UniRef100_A4YNR4 Putative haloacid dehalogenase-like hydrolase, ... 57 1e-06
UniRef100_D0D7B8 HAD-superfamily hydrolase, subfamily IA, varian... 57 1e-06
UniRef100_A5GU30 Putative CbbY homolog (Potential phosphonatase)... 55 3e-06
UniRef100_A5GKS3 Putative CbbY homolog (Potential phosphonatase)... 54 7e-06