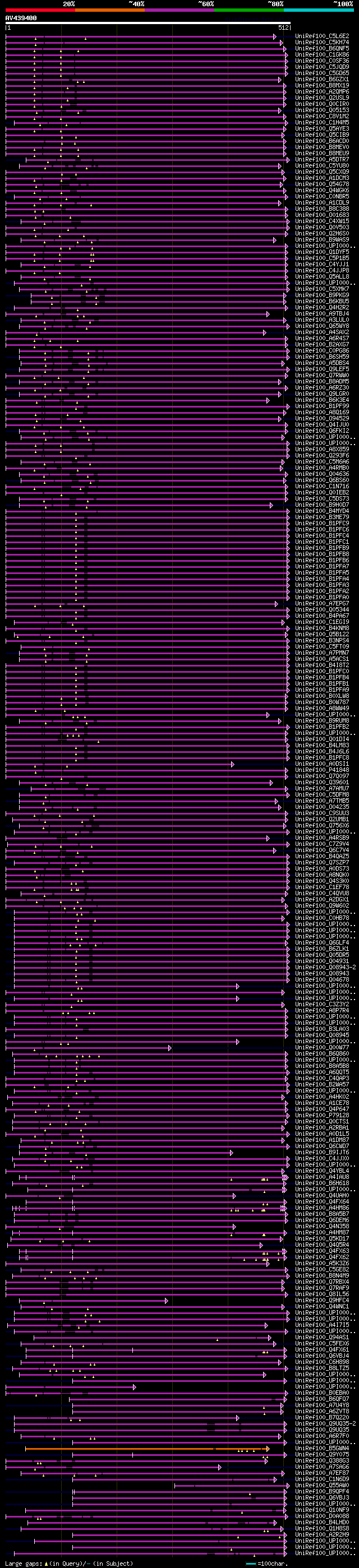
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV439400 PS050a06_r
(512 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_C5L6E2 Structure-specific recognition protein, putativ... 98 3e-19
UniRef100_C5KH74 Structure-specific recognition protein, putativ... 97 5e-19
UniRef100_B6QNF5 Structure-specific recognition protein, putativ... 87 9e-16
UniRef100_C1GK86 FACT complex subunit POB3 n=1 Tax=Paracoccidioi... 86 1e-15
UniRef100_C0SF36 FACT complex subunit pob3 n=1 Tax=Paracoccidioi... 86 1e-15
UniRef100_C5JQD9 FACT complex subunit pob3 n=1 Tax=Ajellomyces d... 85 2e-15
UniRef100_C5GD65 FACT complex subunit pob3 n=1 Tax=Ajellomyces d... 85 2e-15
UniRef100_B6GZX1 Pc12g05980 protein n=1 Tax=Penicillium chrysoge... 85 3e-15
UniRef100_B8MX19 Structure-specific recognition protein, putativ... 84 6e-15
UniRef100_A2QMP6 Function: Pob3 of S. cerevisiae is an essential... 84 6e-15
UniRef100_Q2USL9 FACT complex subunit pob3 n=1 Tax=Aspergillus o... 84 6e-15
UniRef100_Q0CIR0 Putative uncharacterized protein n=1 Tax=Asperg... 84 7e-15
UniRef100_Q05153 FACT complex subunit SSRP1 n=1 Tax=Arabidopsis ... 84 7e-15
UniRef100_C8V1M2 FACT complex subunit pob3 (Facilitates chromati... 83 9e-15
UniRef100_C1H4M5 FACT complex subunit pob3 n=1 Tax=Paracoccidioi... 83 9e-15
UniRef100_Q5AYE3 FACT complex subunit pob3 n=1 Tax=Emericella ni... 83 9e-15
UniRef100_Q5CIB9 Structure specific recognition protein n=1 Tax=... 83 1e-14
UniRef100_B6ACD0 FACT complex subunit SSRP1, putative n=1 Tax=Cr... 82 2e-14
UniRef100_B8MEV0 Structure-specific recognition protein, putativ... 82 3e-14
UniRef100_B8MEU9 Structure-specific recognition protein, putativ... 82 3e-14
UniRef100_A5DTR7 Putative uncharacterized protein n=1 Tax=Lodder... 81 5e-14
UniRef100_C5YU80 Putative uncharacterized protein Sb09g005650 n=... 80 8e-14
UniRef100_Q5CXQ9 Structure-specific recognition protein 1 (SSRP1... 80 8e-14
UniRef100_A1DCM3 Structure-specific recognition protein, putativ... 80 8e-14
UniRef100_Q54G78 FACT complex subunit SSRP1 n=1 Tax=Dictyosteliu... 80 8e-14
UniRef100_Q4WGK6 FACT complex subunit pob3 n=2 Tax=Aspergillus f... 80 8e-14
UniRef100_C0NBR5 FACT complex subunit pob3 n=1 Tax=Ajellomyces c... 80 1e-13
UniRef100_A1CDL9 Structure-specific recognition protein, putativ... 80 1e-13
UniRef100_B8C388 Structure specific recognition protein 1 n=1 Ta... 79 1e-13
UniRef100_O01683 FACT complex subunit ssrp1-B n=1 Tax=Caenorhabd... 79 2e-13
UniRef100_C4XW15 Putative uncharacterized protein n=1 Tax=Clavis... 79 2e-13
UniRef100_Q0V503 Putative uncharacterized protein n=1 Tax=Phaeos... 78 3e-13
UniRef100_Q2H6S0 Putative uncharacterized protein n=1 Tax=Chaeto... 78 4e-13
UniRef100_B9WAS9 FACT complex subunit, putative (Facilitates chr... 78 4e-13
UniRef100_UPI0000E461C4 PREDICTED: similar to HMG box (bp. 1499.... 77 5e-13
UniRef100_Q1DYF5 Putative uncharacterized protein n=1 Tax=Coccid... 77 7e-13
UniRef100_C5P1B5 Structure-specific recognition protein n=1 Tax=... 77 7e-13
UniRef100_C4YJJ1 Putative uncharacterized protein n=1 Tax=Candid... 77 7e-13
UniRef100_C4JJP8 Putative uncharacterized protein n=1 Tax=Uncino... 77 7e-13
UniRef100_Q5ALL8 FACT complex subunit POB3 n=1 Tax=Candida albic... 77 7e-13
UniRef100_UPI00015B6175 PREDICTED: similar to structure-specific... 76 1e-12
UniRef100_C5XMK7 Putative uncharacterized protein Sb03g003450 n=... 76 1e-12
UniRef100_B9PKG9 Structure specific recognition protein, putativ... 76 1e-12
UniRef100_B6KBU5 Structure specific recognition protein I, putat... 76 1e-12
UniRef100_Q4H2R2 FACT complex subunit SSRP1 n=1 Tax=Ciona intest... 76 1e-12
UniRef100_A9TBJ4 Predicted protein (Fragment) n=1 Tax=Physcomitr... 76 2e-12
UniRef100_A3LUL0 DNA polymerase delta binding protein n=1 Tax=Pi... 76 2e-12
UniRef100_Q65WY8 FACT complex subunit SSRP1-B n=3 Tax=Oryza sati... 76 2e-12
UniRef100_A4SAX2 Predicted protein n=1 Tax=Ostreococcus lucimari... 75 2e-12
UniRef100_A6R4S7 Putative uncharacterized protein n=1 Tax=Ajello... 75 2e-12
UniRef100_B2AXG7 Predicted CDS Pa_7_10500 n=1 Tax=Podospora anse... 75 3e-12
UniRef100_C0PG86 Putative uncharacterized protein n=1 Tax=Zea ma... 75 3e-12
UniRef100_B6SH59 Structure-specific recognition protein 1 n=1 Ta... 75 3e-12
UniRef100_A5DBS4 Putative uncharacterized protein n=1 Tax=Pichia... 75 3e-12
UniRef100_Q9LEF5 FACT complex subunit SSRP1 n=1 Tax=Zea mays Rep... 75 3e-12
UniRef100_Q7RWW0 FACT complex subunit pob-3 n=1 Tax=Neurospora c... 75 3e-12
UniRef100_B8ADM5 Putative uncharacterized protein n=1 Tax=Oryza ... 74 4e-12
UniRef100_A6RZ30 Putative uncharacterized protein n=1 Tax=Botryo... 74 4e-12
UniRef100_Q9LGR0 FACT complex subunit SSRP1-A n=2 Tax=Oryza sati... 74 4e-12
UniRef100_B6K3E4 FACT complex subunit pob3 n=1 Tax=Schizosacchar... 74 6e-12
UniRef100_B1PF99 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 73 1e-11
UniRef100_A8Q169 Putative uncharacterized protein n=1 Tax=Malass... 73 1e-11
UniRef100_O94529 FACT complex subunit pob3 n=1 Tax=Schizosacchar... 73 1e-11
UniRef100_Q4IJU0 FACT complex subunit POB3 n=1 Tax=Gibberella ze... 72 2e-11
UniRef100_Q6FKI2 FACT complex subunit POB3 n=1 Tax=Candida glabr... 72 2e-11
UniRef100_UPI000151AAB6 hypothetical protein PGUG_00729 n=1 Tax=... 72 2e-11
UniRef100_UPI0000122EFE hypothetical protein CBG09136 n=1 Tax=Ca... 72 2e-11
UniRef100_A8X859 C. briggsae CBR-HMG-4 protein n=1 Tax=Caenorhab... 72 2e-11
UniRef100_Q293F6 FACT complex subunit Ssrp1 n=2 Tax=pseudoobscur... 72 2e-11
UniRef100_C5M6A6 Putative uncharacterized protein n=1 Tax=Candid... 71 4e-11
UniRef100_A4RMB0 Putative uncharacterized protein n=1 Tax=Magnap... 71 4e-11
UniRef100_Q04636 FACT complex subunit POB3 n=6 Tax=Saccharomyces... 71 4e-11
UniRef100_Q6BS60 FACT complex subunit POB3 n=1 Tax=Debaryomyces ... 71 4e-11
UniRef100_C1N716 Histone chaperone n=1 Tax=Micromonas pusilla CC... 71 5e-11
UniRef100_Q0IEB2 Structure-specific recognition protein n=1 Tax=... 71 5e-11
UniRef100_C5DS73 ZYRO0B14410p n=1 Tax=Zygosaccharomyces rouxii C... 70 6e-11
UniRef100_B9H0D7 High mobility group family n=1 Tax=Populus tric... 70 8e-11
UniRef100_B4MYD4 GK22092 n=1 Tax=Drosophila willistoni RepID=B4M... 70 8e-11
UniRef100_B3ME79 GF12460 n=1 Tax=Drosophila ananassae RepID=B3ME... 70 8e-11
UniRef100_B1PFC9 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFC6 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFC4 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFC1 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFB9 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFB8 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFB6 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA7 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA5 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA4 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA3 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA2 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_B1PFA0 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 70 8e-11
UniRef100_A7EPG7 Putative uncharacterized protein n=1 Tax=Sclero... 70 8e-11
UniRef100_Q05344 FACT complex subunit Ssrp1 n=2 Tax=Drosophila m... 70 8e-11
UniRef100_B4PA67 GE11532 n=1 Tax=Drosophila yakuba RepID=B4PA67_... 70 1e-10
UniRef100_C1EGI9 Predicted protein n=1 Tax=Micromonas sp. RCC299... 69 1e-10
UniRef100_B4KNM8 GI18750 n=1 Tax=Drosophila mojavensis RepID=B4K... 69 1e-10
UniRef100_Q5B122 Histone chaperone rtt106 n=1 Tax=Emericella nid... 69 1e-10
UniRef100_B3NPS4 GG19998 n=1 Tax=Drosophila erecta RepID=B3NPS4_... 69 2e-10
UniRef100_C5FT09 FACT complex subunit pob3 n=1 Tax=Microsporum c... 69 2e-10
UniRef100_A7PMN7 Chromosome chr14 scaffold_21, whole genome shot... 69 2e-10
UniRef100_A5ACS1 Putative uncharacterized protein n=1 Tax=Vitis ... 69 2e-10
UniRef100_B4I8T2 GM15512 n=1 Tax=Drosophila sechellia RepID=B4I8... 69 2e-10
UniRef100_B1PFC0 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 69 2e-10
UniRef100_B1PFB4 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 69 2e-10
UniRef100_B1PFB1 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 69 2e-10
UniRef100_B1PFA9 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 69 2e-10
UniRef100_B0XLW8 FACT complex subunit Ssrp1 n=1 Tax=Culex quinqu... 69 2e-10
UniRef100_B0W787 FACT complex subunit Ssrp1 n=1 Tax=Culex quinqu... 69 2e-10
UniRef100_A8WW49 Putative uncharacterized protein n=1 Tax=Caenor... 69 2e-10
UniRef100_UPI00019274AE PREDICTED: similar to structure specific... 68 3e-10
UniRef100_B9RUM8 Structure-specific recognition protein, putativ... 68 3e-10
UniRef100_B1PFB2 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 68 3e-10
UniRef100_UPI0000D571F7 PREDICTED: similar to AGAP012335-PA n=1 ... 68 4e-10
UniRef100_Q01DI4 Nucleosome-binding factor SPN, POB3 subunit (IS... 68 4e-10
UniRef100_B4LM83 GJ21774 n=1 Tax=Drosophila virilis RepID=B4LM83... 68 4e-10
UniRef100_B4J6L6 GH21151 n=1 Tax=Drosophila grimshawi RepID=B4J6... 68 4e-10
UniRef100_B1PFC8 CG4797 (Fragment) n=1 Tax=Drosophila melanogast... 68 4e-10
UniRef100_A0DSI1 Chromosome undetermined scaffold_61, whole geno... 68 4e-10
UniRef100_P41848 FACT complex subunit SSRP1-A n=1 Tax=Caenorhabd... 67 5e-10
UniRef100_Q7Q097 AGAP012335-PA n=1 Tax=Anopheles gambiae RepID=Q... 67 7e-10
UniRef100_Q39601 FACT complex subunit SSRP1 n=1 Tax=Catharanthus... 67 7e-10
UniRef100_A7AMU7 Structure specific recognition protein, putativ... 67 9e-10
UniRef100_C5DFM8 KLTH0D16390p n=1 Tax=Lachancea thermotolerans C... 67 9e-10
UniRef100_A7TMB5 Putative uncharacterized protein n=1 Tax=Vander... 67 9e-10
UniRef100_O04235 FACT complex subunit SSRP1 n=1 Tax=Vicia faba R... 67 9e-10
UniRef100_C9SUU3 FACT complex subunit pob-3 n=1 Tax=Verticillium... 66 1e-09
UniRef100_Q2UMB1 Histone chaperone rtt106 n=1 Tax=Aspergillus or... 66 1e-09
UniRef100_Q756X6 FACT complex subunit POB3 n=1 Tax=Eremothecium ... 66 1e-09
UniRef100_UPI0001A2CE51 structure specific recognition protein 1... 66 2e-09
UniRef100_A4RSB9 Predicted protein n=1 Tax=Ostreococcus lucimari... 66 2e-09
UniRef100_C7Z9V4 Predicted protein n=1 Tax=Nectria haematococca ... 66 2e-09
UniRef100_Q6C7V4 FACT complex subunit POB3 n=1 Tax=Yarrowia lipo... 66 2e-09
UniRef100_B4QAZ5 GD25013 n=1 Tax=Drosophila simulans RepID=B4QAZ... 65 2e-09
UniRef100_Q7SZP7 Structure specific recognition protein 1a n=1 T... 65 3e-09
UniRef100_A0DS73 Chromosome undetermined scaffold_61, whole geno... 65 3e-09
UniRef100_A8NQK0 Putative uncharacterized protein n=1 Tax=Coprin... 65 3e-09
UniRef100_Q4S3K0 Chromosome 1 SCAF14749, whole genome shotgun se... 65 3e-09
UniRef100_C1EF78 Histone chaperone n=1 Tax=Micromonas sp. RCC299... 65 3e-09
UniRef100_C4QVU8 Subunit of the heterodimeric FACT complex (Spt1... 65 3e-09
UniRef100_A2DGX1 Putative uncharacterized protein n=1 Tax=Tricho... 64 5e-09
UniRef100_Q9W602 FACT complex subunit SSRP1 n=1 Tax=Xenopus laev... 64 5e-09
UniRef100_UPI00017B42A3 UPI00017B42A3 related cluster n=1 Tax=Te... 64 8e-09
UniRef100_C0HB78 FACT complex subunit SSRP1 n=1 Tax=Salmo salar ... 64 8e-09
UniRef100_UPI0001B7B15C FACT complex subunit SSRP1 (Facilitates ... 63 1e-08
UniRef100_UPI00015DF4F4 structure specific recognition protein 1... 63 1e-08
UniRef100_UPI00016E3861 UPI00016E3861 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_Q6GLF4 Ssrp1 protein (Fragment) n=1 Tax=Xenopus (Silur... 63 1e-08
UniRef100_B6ZLK1 Structure-specific recognition protein 1 n=1 Ta... 63 1e-08
UniRef100_Q05DR5 Ssrp1 protein (Fragment) n=2 Tax=Mus musculus R... 63 1e-08
UniRef100_Q04931 FACT complex subunit SSRP1 n=1 Tax=Rattus norve... 63 1e-08
UniRef100_Q08943-2 Isoform 2 of FACT complex subunit SSRP1 n=1 T... 63 1e-08
UniRef100_Q08943 FACT complex subunit SSRP1 n=2 Tax=Mus musculus... 63 1e-08
UniRef100_Q04678 FACT complex subunit SSRP1 n=1 Tax=Gallus gallu... 63 1e-08
UniRef100_UPI0001AFB476 structure specific recognition protein-l... 63 1e-08
UniRef100_UPI0001866958 hypothetical protein BRAFLDRAFT_154810 n... 63 1e-08
UniRef100_UPI0001792697 PREDICTED: similar to FACT complex subun... 63 1e-08
UniRef100_C3Z3Y2 Putative uncharacterized protein (Fragment) n=1... 63 1e-08
UniRef100_A8P7R4 Structure-specific recognition protein 1, putat... 63 1e-08
UniRef100_UPI0001796D0E PREDICTED: structure specific recognitio... 62 2e-08
UniRef100_UPI00006D5771 PREDICTED: similar to structure specific... 62 2e-08
UniRef100_B3LA03 Structure specific recognition protein,putative... 62 2e-08
UniRef100_Q08945 FACT complex subunit SSRP1 n=1 Tax=Homo sapiens... 62 2e-08
UniRef100_UPI0000DB7B26 PREDICTED: similar to Single-strand reco... 62 2e-08
UniRef100_Q00W77 Recombination signal sequence recognition pr (I... 62 2e-08
UniRef100_B6Q860 Negative regulator of DNA transposition (Rtt106... 62 2e-08
UniRef100_UPI000175F362 PREDICTED: structure specific recognitio... 62 3e-08
UniRef100_B8A5B8 Structure specific recognition protein 1b (Frag... 62 3e-08
UniRef100_A6QQT5 SSRP1 protein n=1 Tax=Bos taurus RepID=A6QQT5_B... 62 3e-08
UniRef100_C4QAP3 Structure specific recognition protein, putativ... 62 3e-08
UniRef100_B2WA57 FACT complex subunit pob3 n=1 Tax=Pyrenophora t... 62 3e-08
UniRef100_UPI0000F2DA9F PREDICTED: similar to high mobility grou... 61 4e-08
UniRef100_A4HK02 Putative uncharacterized protein n=1 Tax=Leishm... 61 4e-08
UniRef100_A1CE78 Histone chaperone rtt106 n=1 Tax=Aspergillus cl... 61 4e-08
UniRef100_Q4P647 FACT complex subunit POB3 n=1 Tax=Ustilago mayd... 61 4e-08
UniRef100_P79128 Structure-specific recognition protein 1 (Fragm... 61 5e-08
UniRef100_Q0CTS1 Histone chaperone rtt106 n=1 Tax=Aspergillus te... 61 5e-08
UniRef100_A2RBA1 Histone chaperone rtt106 n=1 Tax=Aspergillus ni... 61 5e-08
UniRef100_A0D1L5 Chromosome undetermined scaffold_34, whole geno... 60 7e-08
UniRef100_A1DM87 Histone chaperone rtt106 n=1 Tax=Neosartorya fi... 60 7e-08
UniRef100_Q6CWD7 FACT complex subunit POB3 n=1 Tax=Kluyveromyces... 60 7e-08
UniRef100_B9IJT6 High mobility group family n=1 Tax=Populus tric... 60 9e-08
UniRef100_C4JJX0 Putative uncharacterized protein n=1 Tax=Uncino... 60 9e-08
UniRef100_UPI00016E388A UPI00016E388A related cluster n=1 Tax=Ta... 60 1e-07
UniRef100_Q4YBL4 Structure specific recognition protein, putativ... 60 1e-07
UniRef100_A4IAU8 Proteophosphoglycan ppg4 n=1 Tax=Leishmania inf... 60 1e-07
UniRef100_B6H618 Pc14g01900 protein n=1 Tax=Penicillium chrysoge... 60 1e-07
UniRef100_UPI0001760070 PREDICTED: hypothetical protein, partial... 59 1e-07
UniRef100_Q4UAH0 Structure-specific recognition protein (SSRP) 1... 59 1e-07
UniRef100_Q4FX64 Proteophosphoglycan ppg3, putative n=1 Tax=Leis... 59 1e-07
UniRef100_A4HM86 Proteophosphoglycan ppg4 n=1 Tax=Leishmania bra... 59 1e-07
UniRef100_B8A5B7 Structure specific recognition protein 1b (Frag... 59 2e-07
UniRef100_Q6DEM6 Ssrp1b protein (Fragment) n=1 Tax=Danio rerio R... 59 2e-07
UniRef100_Q4N358 Structure specific recognition protein, putativ... 59 2e-07
UniRef100_A4HM87 Proteophosphoglycan ppg4 n=1 Tax=Leishmania bra... 59 2e-07
UniRef100_Q5KD17 FACT complex subunit POB3 n=1 Tax=Filobasidiell... 59 2e-07
UniRef100_Q4Q5R4 Putative uncharacterized protein n=1 Tax=Leishm... 59 2e-07
UniRef100_Q4FX63 Proteophosphoglycan ppg4 n=1 Tax=Leishmania maj... 59 2e-07
UniRef100_Q4FX62 Proteophosphoglycan 5 n=1 Tax=Leishmania major ... 59 2e-07
UniRef100_A5K3Z6 Structure specific recognition protein, putativ... 59 2e-07
UniRef100_C5GE82 Histone chaperone RTT106 n=2 Tax=Ajellomyces de... 59 2e-07
UniRef100_B8N4M9 Negative regulator of DNA transposition (Rtt106... 59 2e-07
UniRef100_Q7RBX4 Putative structure specific recognition protein... 58 3e-07
UniRef100_Q7RAF9 Structure-specific recognition protein 1 (Fragm... 58 3e-07
UniRef100_Q8IL56 Structure specific recognition protein n=1 Tax=... 58 4e-07
UniRef100_Q9HFC4 SSRP1-like protein (Fragment) n=1 Tax=Zygosacch... 58 4e-07
UniRef100_Q4WNC1 Histone chaperone rtt106 n=2 Tax=Aspergillus fu... 58 4e-07
UniRef100_UPI00005A35CE PREDICTED: similar to structure specific... 57 6e-07
UniRef100_UPI00005A35CD PREDICTED: similar to structure specific... 57 6e-07
UniRef100_A4I7I5 Putative uncharacterized protein n=1 Tax=Leishm... 57 6e-07
UniRef100_UPI00005A35CF PREDICTED: similar to structure specific... 57 7e-07
UniRef100_Q94AS1 Putative arginine/serine-rich protein n=1 Tax=A... 57 7e-07
UniRef100_C5FEX6 Meiotically up-regulated gene 183 protein n=1 T... 57 9e-07
UniRef100_Q4FX61 Proteophosphoglycan ppg1 n=2 Tax=Leishmania maj... 56 1e-06
UniRef100_Q6VBJ4 Epa5p n=1 Tax=Candida glabrata RepID=Q6VBJ4_CANGA 56 1e-06
UniRef100_C6H898 Histone chaperone Rttp106-like protein n=1 Tax=... 56 1e-06
UniRef100_B8LTZ5 Negative regulator of DNA transposition (Rtt106... 56 1e-06
UniRef100_UPI000186D023 predicted protein n=1 Tax=Pediculus huma... 56 2e-06
UniRef100_UPI00015537C6 PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000E4A829 PREDICTED: similar to HMG box (bp. 1499.... 56 2e-06
UniRef100_B0EBA0 FACT complex subunit SSRP1-A, putative n=1 Tax=... 56 2e-06
UniRef100_B6QFQ7 Class III chitinase ChiA1 n=1 Tax=Penicillium m... 56 2e-06
UniRef100_A7U4Y8 Flocculin n=1 Tax=Saccharomyces cerevisiae RepI... 56 2e-06
UniRef100_A6ZVT8 Cell surface flocculin n=1 Tax=Saccharomyces ce... 56 2e-06
UniRef100_B7Q220 Structure-specific recognition protein, putativ... 55 2e-06
UniRef100_Q9UQ35-2 Isoform 2 of Serine/arginine repetitive matri... 55 2e-06
UniRef100_Q9UQ35 Serine/arginine repetitive matrix protein 2 n=1... 55 2e-06
UniRef100_A6R7F0 Histone chaperone RTT106 n=1 Tax=Ajellomyces ca... 55 3e-06
UniRef100_UPI00016E1500 UPI00016E1500 related cluster n=1 Tax=Ta... 55 4e-06
UniRef100_B5GWN4 Alpha-galactosidase n=1 Tax=Streptomyces clavul... 55 4e-06
UniRef100_Q9Y075 Proteophosphoglycan (Fragment) n=1 Tax=Leishman... 55 4e-06
UniRef100_Q388G3 Putative uncharacterized protein n=1 Tax=Trypan... 55 4e-06
UniRef100_A7SAG6 Predicted protein (Fragment) n=1 Tax=Nematostel... 55 4e-06
UniRef100_A7EF87 Putative uncharacterized protein n=1 Tax=Sclero... 55 4e-06
UniRef100_C1N6D9 Predicted protein n=1 Tax=Micromonas pusilla CC... 54 5e-06
UniRef100_Q55AW0 3'-5' exonuclease n=1 Tax=Dictyostelium discoid... 54 5e-06
UniRef100_B9QPF4 DNA mismatch repair protein, putative n=1 Tax=T... 54 5e-06
UniRef100_Q6VBJ3 Epa4p n=1 Tax=Candida glabrata RepID=Q6VBJ3_CANGA 54 5e-06
UniRef100_UPI00015539A6 PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_Q10NF9 Retrotransposon protein, putative, unclassified... 54 6e-06
UniRef100_D0A088 Putative uncharacterized protein n=1 Tax=Trypan... 54 6e-06
UniRef100_B4LHD0 GJ12692 n=1 Tax=Drosophila virilis RepID=B4LHD0... 54 6e-06
UniRef100_Q1H8S8 Cell surface flocculin n=1 Tax=Saccharomyces ce... 54 6e-06
UniRef100_A2R2H9 Similarity: the similarities are due to repetet... 54 6e-06
UniRef100_UPI0001551D8C trinucleotide repeat containing 18 n=1 T... 54 8e-06
UniRef100_UPI0000141A8E tetra-peptide repeat homeobox-like (TPRX... 54 8e-06
UniRef100_UPI000179CC90 UPI000179CC90 related cluster n=1 Tax=Bo... 54 8e-06
UniRef100_Q17RH7 Putative protein TPRXL n=1 Tax=Homo sapiens Rep... 54 8e-06