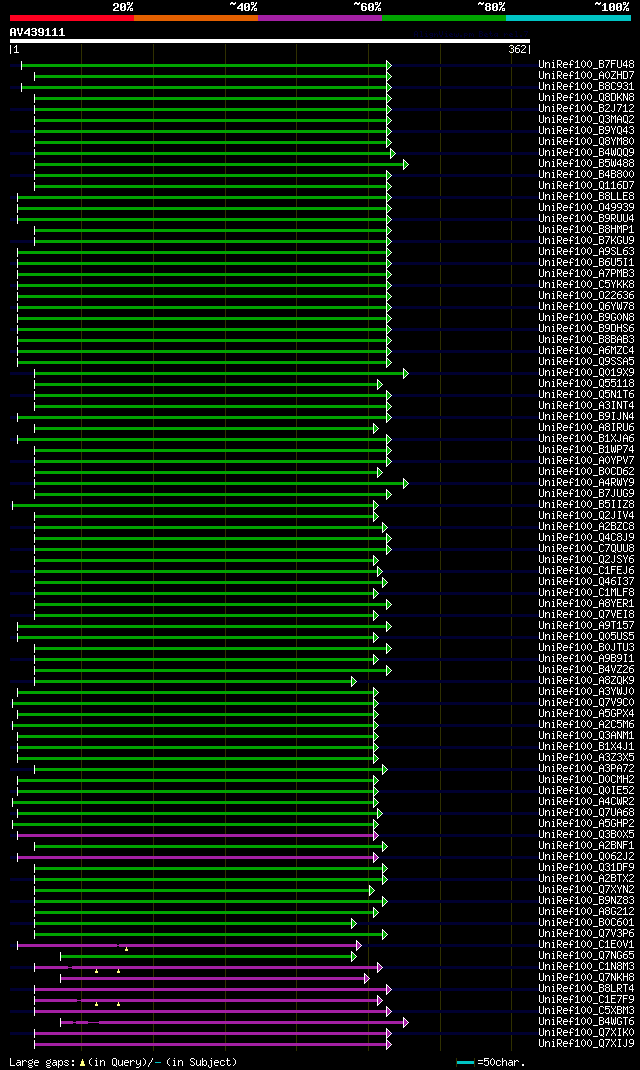
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV439111 PS045g08_r
(362 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B7FU48 Predicted protein n=1 Tax=Phaeodactylum tricorn... 109 1e-22
UniRef100_A0ZHD7 Peptidyl-prolyl cis-trans isomerase, cyclophili... 107 3e-22
UniRef100_B8C931 Predicted protein n=1 Tax=Thalassiosira pseudon... 107 4e-22
UniRef100_Q8DKN8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=The... 107 5e-22
UniRef100_B2J712 Peptidyl-prolyl cis-trans isomerase, cyclophili... 107 5e-22
UniRef100_Q3MAQ2 Peptidyl-prolyl cis-trans isomerase, cyclophili... 106 7e-22
UniRef100_B9YQ43 Peptidyl-prolyl cis-trans isomerase cyclophilin... 106 9e-22
UniRef100_Q8YM80 Alr5059 protein n=1 Tax=Nostoc sp. PCC 7120 Rep... 105 2e-21
UniRef100_B4WQQ9 Peptidyl-prolyl cis-trans isomerase, cyclophili... 105 2e-21
UniRef100_B5W488 Peptidyl-prolyl cis-trans isomerase cyclophilin... 103 5e-21
UniRef100_B4B800 Peptidyl-prolyl cis-trans isomerase cyclophilin... 101 2e-20
UniRef100_Q116D7 Peptidyl-prolyl cis-trans isomerase, cyclophili... 100 5e-20
UniRef100_B8LLE8 Putative uncharacterized protein n=1 Tax=Picea ... 100 5e-20
UniRef100_O49939 Peptidyl-prolyl cis-trans isomerase, chloroplas... 100 7e-20
UniRef100_B9RUU4 Peptidyl-prolyl cis-trans isomerase, putative n... 100 9e-20
UniRef100_B8HMP1 Peptidyl-prolyl cis-trans isomerase cyclophilin... 99 1e-19
UniRef100_B7KGU9 Peptidyl-prolyl cis-trans isomerase cyclophilin... 99 1e-19
UniRef100_A9SL63 Predicted protein n=1 Tax=Physcomitrella patens... 99 1e-19
UniRef100_B6U5I1 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Zea... 98 3e-19
UniRef100_A7PMB3 Chromosome chr14 scaffold_21, whole genome shot... 98 3e-19
UniRef100_C5YKK8 Putative uncharacterized protein Sb07g019320 n=... 98 3e-19
UniRef100_O22636 Poly(A) polymerase n=1 Tax=Pisum sativum RepID=... 97 4e-19
UniRef100_Q6YW78 Os08g0382400 protein n=1 Tax=Oryza sativa Japon... 97 6e-19
UniRef100_B9G0N8 Putative uncharacterized protein n=1 Tax=Oryza ... 97 6e-19
UniRef100_B9DHS6 AT3G01480 protein (Fragment) n=1 Tax=Arabidopsi... 97 6e-19
UniRef100_B8BAB3 Putative uncharacterized protein n=1 Tax=Oryza ... 97 6e-19
UniRef100_A6MZC4 Peptidyl-prolyl cis-trans isomerase (Fragment) ... 97 6e-19
UniRef100_Q9SSA5 Peptidyl-prolyl cis-trans isomerase CYP38, chlo... 97 6e-19
UniRef100_Q019X9 Cyclophilin type, U box-containing peptidyl-pro... 96 1e-18
UniRef100_Q55118 Putative thylakoid lumen peptidyl-prolyl cis-tr... 96 1e-18
UniRef100_Q5N1T6 Peptidyl-prolyl cis-trans isomerase n=3 Tax=Syn... 96 2e-18
UniRef100_A3INT4 Putative uncharacterized protein n=1 Tax=Cyanot... 95 2e-18
UniRef100_B9IJN4 Predicted protein n=1 Tax=Populus trichocarpa R... 95 2e-18
UniRef100_A8IRU6 Peptidyl-prolyl cis-trans isomerase, cyclophili... 95 2e-18
UniRef100_B1XJA6 Putative peptidyl-prolyl cis-trans isomerase n=... 95 3e-18
UniRef100_B1WP74 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 94 4e-18
UniRef100_A0YPV7 Peptidyl-prolyl cis-trans isomerase, cyclophili... 94 4e-18
UniRef100_B0CD62 Peptidyl-prolyl cis-trans isomerase, cyclophili... 93 1e-17
UniRef100_A4RWY9 Predicted protein n=1 Tax=Ostreococcus lucimari... 93 1e-17
UniRef100_B7JUG9 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Cya... 92 1e-17
UniRef100_B5IIZ8 Peptidyl-prolyl cis-trans isomerase, cyclophili... 92 2e-17
UniRef100_Q2JIV4 Peptidyl-prolyl cis-trans isomerase, cyclophili... 91 3e-17
UniRef100_A2BZC8 Cyclophilin-type peptidyl-prolyl cis-trans isom... 91 5e-17
UniRef100_Q4C8J9 Putative uncharacterized protein n=1 Tax=Crocos... 91 5e-17
UniRef100_C7QUU8 Peptidylprolyl isomerase n=1 Tax=Cyanothece sp.... 91 5e-17
UniRef100_Q2JSY6 Peptidyl-prolyl cis-trans isomerase, cyclophili... 90 7e-17
UniRef100_C1FEJ6 Peptidyl-prolyl cis-trans isomerase chloroplast... 90 7e-17
UniRef100_Q46I37 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pro... 90 9e-17
UniRef100_C1MLF8 Peptidyl-prolyl cis-trans isomerase chloroplast... 90 9e-17
UniRef100_A8YER1 Genome sequencing data, contig C300 n=1 Tax=Mic... 89 2e-16
UniRef100_Q7VEI8 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Pro... 89 2e-16
UniRef100_A9T157 Predicted protein n=1 Tax=Physcomitrella patens... 88 3e-16
UniRef100_Q05US5 Putative cyclophilin-type peptidyl-prolyl cis-t... 87 6e-16
UniRef100_B0JTU3 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Mic... 87 8e-16
UniRef100_A9B9I1 Cyclophilin-type peptidyl-prolyl cis-trans isom... 87 8e-16
UniRef100_B4VZ26 Peptidyl-prolyl cis-trans isomerase, cyclophili... 86 1e-15
UniRef100_A8ZQK9 Peptidyl-prolyl cis-trans isomerase, cyclophili... 85 2e-15
UniRef100_A3YWJ0 Putative cyclophilin-type peptidyl-prolyl cis-t... 84 4e-15
UniRef100_Q7V9C0 Cyclophilin-type peptidyl-prolyl cis-trans isom... 84 5e-15
UniRef100_A5GPX4 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Syn... 84 5e-15
UniRef100_A2C5M6 Cyclophilin-type peptidyl-prolyl cis-trans isom... 84 5e-15
UniRef100_Q3ANM1 Putative cyclophilin-type peptidyl-prolyl cis-t... 83 8e-15
UniRef100_B1X4J1 Putative cyclophilin-type peptidyl-prolyl cis-t... 82 1e-14
UniRef100_A3Z3X5 Putative cyclophilin-type peptidyl-prolyl cis-t... 82 2e-14
UniRef100_A3PA72 Cyclophilin-type peptidyl-prolyl cis-trans isom... 81 3e-14
UniRef100_D0CMH2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Syn... 81 3e-14
UniRef100_Q0IE52 Peptidyl-prolyl cis-trans isomerase, cyclophili... 80 5e-14
UniRef100_A4CWR2 Putative cyclophilin-type peptidyl-prolyl cis-t... 80 9e-14
UniRef100_Q7UA68 Putative cyclophilin-type peptidyl-prolyl cis-t... 79 1e-13
UniRef100_A5GHP2 Peptidyl-prolyl cis-trans isomerase n=1 Tax=Syn... 79 2e-13
UniRef100_Q3B0X5 Putative cyclophilin-type peptidyl-prolyl cis-t... 79 2e-13
UniRef100_A2BNF1 Cyclophilin-type peptidyl-prolyl cis-trans isom... 79 2e-13
UniRef100_Q062J2 Putative cyclophilin-type peptidyl-prolyl cis-t... 79 2e-13
UniRef100_Q31DF9 Cyclophilin-type peptidyl-prolyl cis-trans isom... 78 3e-13
UniRef100_A2BTX2 Cyclophilin-type peptidyl-prolyl cis-trans isom... 78 3e-13
UniRef100_Q7XYN2 Thylakoid lumen protease (Fragment) n=1 Tax=Big... 78 3e-13
UniRef100_B9NZ83 Cyclophilin-type peptidyl-prolyl cis-trans isom... 78 3e-13
UniRef100_A8G212 Cyclophilin-type peptidyl-prolyl cis-trans isom... 77 6e-13
UniRef100_B0C601 Peptidyl-prolyl cis-trans isomerase, cyclophili... 75 3e-12
UniRef100_Q7V3P6 Cyclophilin-type peptidyl-prolyl cis-trans isom... 73 9e-12
UniRef100_C1E0V1 Predicted protein n=1 Tax=Micromonas sp. RCC299... 63 1e-08
UniRef100_Q7NG65 Gll3308 protein n=1 Tax=Gloeobacter violaceus R... 60 6e-08
UniRef100_C1N8M3 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 1e-07
UniRef100_Q7NKH8 Glr1500 protein n=1 Tax=Gloeobacter violaceus R... 58 4e-07
UniRef100_B8LRT4 Putative uncharacterized protein n=1 Tax=Picea ... 56 1e-06
UniRef100_C1E7F9 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 2e-06
UniRef100_C5XBM3 Putative uncharacterized protein Sb02g036560 n=... 55 3e-06
UniRef100_B4WGT6 Peptidyl-prolyl cis-trans isomerase, cyclophili... 54 4e-06
UniRef100_Q7XIK0 Putative peptidyl-prolycis-trans isomerase prot... 53 9e-06
UniRef100_Q7XIJ9 Os07g0565600 protein n=1 Tax=Oryza sativa Japon... 53 9e-06
UniRef100_B8B7H0 Putative uncharacterized protein n=1 Tax=Oryza ... 53 9e-06