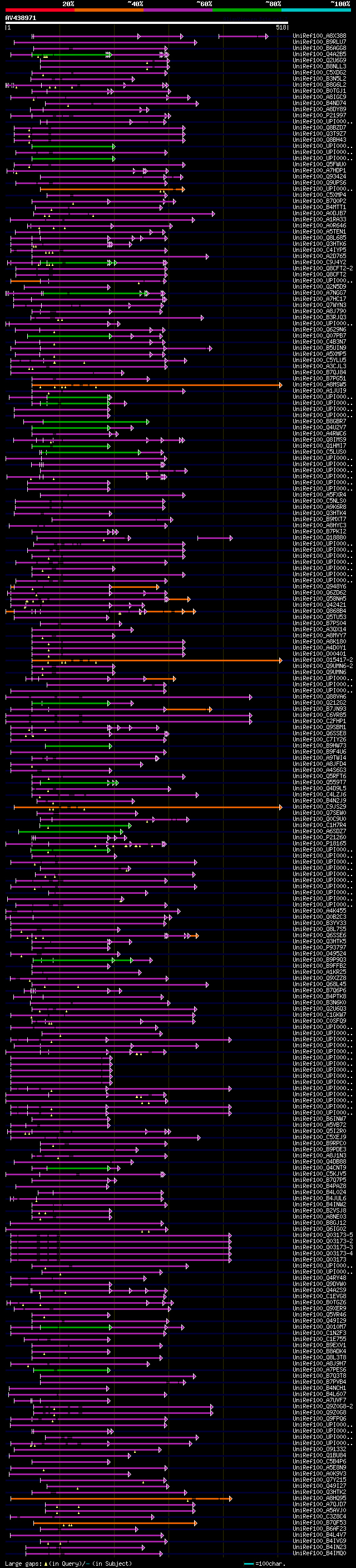
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV438971 PS043h08_r
(518 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_A8X388 C. briggsae CBR-GRL-25 protein n=1 Tax=Caenorha... 64 5e-10
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 65 2e-09
UniRef100_B6AGG8 Putative uncharacterized protein n=1 Tax=Crypto... 64 8e-09
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 62 2e-08
UniRef100_Q2U6G9 Predicted protein n=1 Tax=Aspergillus oryzae Re... 62 2e-08
UniRef100_B8NLL3 Putative uncharacterized protein n=1 Tax=Asperg... 62 2e-08
UniRef100_C5XDG2 Putative uncharacterized protein Sb02g006780 n=... 62 2e-08
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 62 2e-08
UniRef100_B8G6L2 Putative uncharacterized protein n=1 Tax=Chloro... 62 3e-08
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 62 3e-08
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 62 3e-08
UniRef100_B4ND74 GK24962 n=1 Tax=Drosophila willistoni RepID=B4N... 62 3e-08
UniRef100_A8DY89 CG34220 n=1 Tax=Drosophila melanogaster RepID=A... 62 3e-08
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 61 4e-08
UniRef100_UPI0000E491FD PREDICTED: hypothetical protein, partial... 61 5e-08
UniRef100_Q8BZD7 Putative uncharacterized protein (Fragment) n=2... 61 5e-08
UniRef100_Q3T9Z7 Putative uncharacterized protein n=1 Tax=Mus mu... 61 5e-08
UniRef100_Q8BH43 Wiskott-Aldrich syndrome protein family member ... 61 5e-08
UniRef100_UPI0000EBE712 PREDICTED: similar to MYST histone acety... 60 7e-08
UniRef100_UPI0001AE6B4C SET domain-containing protein 1B n=1 Tax... 60 7e-08
UniRef100_UPI000179CEFB UPI000179CEFB related cluster n=1 Tax=Bo... 60 7e-08
UniRef100_Q5FWU0 WAS protein family, member 2 n=1 Tax=Rattus nor... 60 7e-08
UniRef100_A7HDP1 SpoIID/LytB domain n=1 Tax=Anaeromyxobacter sp.... 60 7e-08
UniRef100_Q93424 Protein E02A10.2, partially confirmed by transc... 60 7e-08
UniRef100_Q9UPS6 Histone-lysine N-methyltransferase SETD1B n=1 T... 60 7e-08
UniRef100_UPI0000222A03 Hypothetical protein CBG23530 n=1 Tax=Ca... 60 9e-08
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 60 9e-08
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 60 9e-08
UniRef100_B4MTT1 GK23771 n=1 Tax=Drosophila willistoni RepID=B4M... 60 9e-08
UniRef100_A0DJB7 Chromosome undetermined scaffold_53, whole geno... 60 9e-08
UniRef100_A1RA33 Putative lipoprotein n=1 Tax=Arthrobacter aures... 60 1e-07
UniRef100_A0R646 Transcriptional regulator n=1 Tax=Mycobacterium... 60 1e-07
UniRef100_A5TEN1 Putative polyketide synthase n=1 Tax=Burkholder... 60 1e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 60 1e-07
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 60 1e-07
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 60 1e-07
UniRef100_A2D765 Putative uncharacterized protein n=1 Tax=Tricho... 60 1e-07
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 60 1e-07
UniRef100_Q8CFT2-2 Isoform 2 of Histone-lysine N-methyltransfera... 60 1e-07
UniRef100_Q8CFT2 Histone-lysine N-methyltransferase SETD1B n=2 T... 60 1e-07
UniRef100_UPI0000F2BD8B PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 59 1e-07
UniRef100_A7NGG7 Integrin alpha beta-propellor repeat protein n=... 59 1e-07
UniRef100_A7HC17 WD40 domain protein beta Propeller n=1 Tax=Anae... 59 1e-07
UniRef100_Q7WYN3 Cellulosomal scaffoldin adaptor protein B n=1 T... 59 1e-07
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 59 1e-07
UniRef100_B3RJQ3 Putative uncharacterized protein n=1 Tax=Tricho... 59 1e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 59 2e-07
UniRef100_Q629N6 Putative polyketide synthase n=1 Tax=Burkholder... 59 2e-07
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 59 2e-07
UniRef100_C4B3N7 Multi-domain beta keto-acyl synthase n=1 Tax=Bu... 59 2e-07
UniRef100_B5UIN9 Surface protein n=1 Tax=Bacillus cereus AH1134 ... 59 2e-07
UniRef100_A5XMP5 Putative polyketide synthase n=1 Tax=Burkholder... 59 2e-07
UniRef100_C5YLU5 Putative uncharacterized protein Sb07g000890 n=... 59 2e-07
UniRef100_A3CJL3 Putative uncharacterized protein n=1 Tax=Oryza ... 59 2e-07
UniRef100_B7QJ84 Glycine-rich RNA-binding protein GRP1A, putativ... 59 2e-07
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 59 2e-07
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 59 2e-07
UniRef100_A1JUI9 WASL protein n=1 Tax=Homo sapiens RepID=A1JUI9_... 59 2e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 59 3e-07
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 59 3e-07
UniRef100_B8GBR7 Lipolytic protein G-D-S-L family n=1 Tax=Chloro... 59 3e-07
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 59 3e-07
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 59 3e-07
UniRef100_Q8IMS9 CG31439 n=1 Tax=Drosophila melanogaster RepID=Q... 59 3e-07
UniRef100_Q1HMI7 Formin B n=2 Tax=Trypanosoma cruzi RepID=Q1HMI7... 59 3e-07
UniRef100_C5LUS0 Merozoite surface protein 2, putative (Fragment... 59 3e-07
UniRef100_UPI000176147B PREDICTED: hypothetical protein, partial... 58 3e-07
UniRef100_UPI0000E48F2E PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI0000E48095 PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI0000DA3A96 PREDICTED: hypothetical protein n=1 Tax=... 58 3e-07
UniRef100_UPI00016E10B6 UPI00016E10B6 related cluster n=1 Tax=Ta... 58 3e-07
UniRef100_UPI00016E10A1 UPI00016E10A1 related cluster n=1 Tax=Ta... 58 3e-07
UniRef100_A5FXR4 TonB family protein n=1 Tax=Acidiphilium cryptu... 58 3e-07
UniRef100_C5NLS0 Multi-domain beta keto-acyl synthase n=1 Tax=Bu... 58 3e-07
UniRef100_A9K6R8 Putative polyketide synthase n=1 Tax=Burkholder... 58 3e-07
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 58 3e-07
UniRef100_B9MXT7 Predicted protein n=1 Tax=Populus trichocarpa R... 58 3e-07
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 58 3e-07
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 58 3e-07
UniRef100_Q18880 Protein C56A3.1, partially confirmed by transcr... 53 4e-07
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 58 4e-07
UniRef100_UPI000194D9A6 PREDICTED: similar to WAS protein family... 58 4e-07
UniRef100_UPI0001818C5C Wiskott-Aldrich syndrome-like n=1 Tax=Po... 58 4e-07
UniRef100_UPI00017C32F4 PREDICTED: similar to Histone-lysine N-m... 58 4e-07
UniRef100_UPI0000E25181 PREDICTED: myeloid/lymphoid or mixed-lin... 58 4e-07
UniRef100_UPI0000E2171E PREDICTED: Wiskott-Aldrich syndrome gene... 58 4e-07
UniRef100_UPI000179DAA8 UPI000179DAA8 related cluster n=1 Tax=Bo... 58 4e-07
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 58 4e-07
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 58 4e-07
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 58 4e-07
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 58 4e-07
UniRef100_Q868B4 Protein ZK643.8, partially confirmed by transcr... 58 4e-07
UniRef100_Q5TU53 AGAP002965-PA (Fragment) n=1 Tax=Anopheles gamb... 58 4e-07
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 58 4e-07
UniRef100_A3QX14 Wiskott-Aldrich syndrome protein (Fragment) n=1... 58 4e-07
UniRef100_A8MVY7 Putative uncharacterized protein ENSP0000038013... 58 4e-07
UniRef100_A8K180 cDNA FLJ76749, highly similar to Homo sapiens W... 58 4e-07
UniRef100_A4D0Y1 Wiskott-Aldrich syndrome-like n=1 Tax=Homo sapi... 58 4e-07
UniRef100_O00401 Neural Wiskott-Aldrich syndrome protein n=1 Tax... 58 4e-07
UniRef100_O15417-2 Isoform 2 of Trinucleotide repeat-containing ... 58 4e-07
UniRef100_Q9UMN6-2 Isoform 2 of Histone-lysine N-methyltransfera... 58 4e-07
UniRef100_Q9UMN6 Histone-lysine N-methyltransferase MLL4 n=1 Tax... 58 4e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 57 6e-07
UniRef100_UPI0000E47DFA PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_UPI0000DF063E Os02g0254800 n=1 Tax=Oryza sativa Japoni... 57 6e-07
UniRef100_Q88VA6 Extracellular protein, gamma-D-glutamate-meso-d... 57 6e-07
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 57 6e-07
UniRef100_B7JN93 Lpxtg-motif cell wall anchor domain protein n=1... 57 6e-07
UniRef100_C6VR85 Extracellular protein, gamma-D-glutamate-meso-d... 57 6e-07
UniRef100_C2FHP1 Extracellular protein, gamma-D-glutamate-meso-d... 57 6e-07
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 57 6e-07
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 57 6e-07
UniRef100_C7IY26 Os02g0254800 protein n=1 Tax=Oryza sativa Japon... 57 6e-07
UniRef100_B9HW73 Predicted protein n=1 Tax=Populus trichocarpa R... 57 6e-07
UniRef100_B9F4U6 Putative uncharacterized protein n=1 Tax=Oryza ... 57 6e-07
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 57 6e-07
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 57 6e-07
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 57 6e-07
UniRef100_Q5RFT6 Putative uncharacterized protein DKFZp459G071 n... 57 6e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 57 6e-07
UniRef100_Q4D9L5 Putative uncharacterized protein n=1 Tax=Trypan... 57 6e-07
UniRef100_C4LZJ6 Putative uncharacterized protein n=1 Tax=Entamo... 57 6e-07
UniRef100_B4N2J9 GK19249 n=1 Tax=Drosophila willistoni RepID=B4N... 57 6e-07
UniRef100_C9JS29 Putative uncharacterized protein TNRC18 n=1 Tax... 57 6e-07
UniRef100_Q7SEW0 Predicted protein n=1 Tax=Neurospora crassa Rep... 57 6e-07
UniRef100_Q0C9U0 Putative uncharacterized protein n=1 Tax=Asperg... 57 6e-07
UniRef100_C1H7R4 Putative uncharacterized protein n=1 Tax=Paraco... 57 6e-07
UniRef100_A6SDZ7 Predicted protein n=1 Tax=Botryotinia fuckelian... 57 6e-07
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 57 6e-07
UniRef100_P18165 Loricrin n=1 Tax=Mus musculus RepID=LORI_MOUSE 57 6e-07
UniRef100_UPI0001A7B3C7 unknown protein n=1 Tax=Arabidopsis thal... 57 7e-07
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 57 7e-07
UniRef100_UPI00017600E2 PREDICTED: im:7158925 n=1 Tax=Danio reri... 57 7e-07
UniRef100_UPI0000E48C90 PREDICTED: hypothetical protein n=1 Tax=... 57 7e-07
UniRef100_UPI0000E4896A PREDICTED: similar to CG33556-PA n=1 Tax... 57 7e-07
UniRef100_UPI00005A499E PREDICTED: similar to CG40351-PA.3 n=1 T... 57 7e-07
UniRef100_UPI0001A2C389 UPI0001A2C389 related cluster n=1 Tax=Da... 57 7e-07
UniRef100_UPI0000DC131C UPI0000DC131C related cluster n=1 Tax=Ra... 57 7e-07
UniRef100_UPI00016E37F0 UPI00016E37F0 related cluster n=1 Tax=Ta... 57 7e-07
UniRef100_UPI000184A044 SET domain-containing protein 1B n=1 Tax... 57 7e-07
UniRef100_A4K455 Antifreeze glycoprotein n=1 Tax=Boreogadus said... 57 7e-07
UniRef100_Q0B2C3 Putative uncharacterized protein n=1 Tax=Burkho... 57 7e-07
UniRef100_B3YV33 Surface protein, pentapeptide repeat domain n=1... 57 7e-07
UniRef100_Q8L7S5 AT4g18560/F28J12_220 n=1 Tax=Arabidopsis thalia... 57 7e-07
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 57 7e-07
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 57 7e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 57 7e-07
UniRef100_O49524 Pherophorin - like protein n=1 Tax=Arabidopsis ... 57 7e-07
UniRef100_B9P9Q3 Predicted protein (Fragment) n=1 Tax=Populus tr... 57 7e-07
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 57 7e-07
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 57 7e-07
UniRef100_Q9XZZ8 Secretory protein (LS110p) n=1 Tax=Litomosoides... 57 7e-07
UniRef100_Q68L45 Optomotor blind (Fragment) n=1 Tax=Drosophila p... 57 7e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 57 7e-07
UniRef100_B4PTK8 GE10695 n=1 Tax=Drosophila yakuba RepID=B4PTK8_... 57 7e-07
UniRef100_B3N6K0 GG25239 n=1 Tax=Drosophila erecta RepID=B3N6K0_... 57 7e-07
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 57 7e-07
UniRef100_C1GKW7 Predicted protein n=1 Tax=Paracoccidioides bras... 57 7e-07
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 57 7e-07
UniRef100_UPI0001791EEC PREDICTED: similar to CG31374 CG31374-PB... 57 1e-06
UniRef100_UPI000155EC14 PREDICTED: similar to SET binding protei... 57 1e-06
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 57 1e-06
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 57 1e-06
UniRef100_UPI0000DA1F7B PREDICTED: similar to Loricrin n=1 Tax=R... 57 1e-06
UniRef100_UPI0000D9CC90 PREDICTED: similar to keratin 1 isoform ... 57 1e-06
UniRef100_UPI0000D9CC8F PREDICTED: similar to keratin 1 isoform ... 57 1e-06
UniRef100_UPI0000D9CC8E PREDICTED: similar to keratin 1 isoform ... 57 1e-06
UniRef100_UPI0000D9CC8D PREDICTED: similar to keratin 1 isoform ... 57 1e-06
UniRef100_UPI0000D9CC8C PREDICTED: similar to keratin 1 isoform ... 57 1e-06
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 57 1e-06
UniRef100_UPI0000DC09EF UPI0000DC09EF related cluster n=1 Tax=Ra... 57 1e-06
UniRef100_UPI0000504055 Keratin, type II cytoskeletal 2 epiderma... 57 1e-06
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 57 1e-06
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 57 1e-06
UniRef100_B6INW7 R body protein, putative n=1 Tax=Rhodospirillum... 57 1e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 57 1e-06
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 57 1e-06
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 57 1e-06
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 57 1e-06
UniRef100_B9PDE3 Predicted protein (Fragment) n=1 Tax=Populus tr... 57 1e-06
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 57 1e-06
UniRef100_Q4DB88 Putative uncharacterized protein n=1 Tax=Trypan... 57 1e-06
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 57 1e-06
UniRef100_C5KJV5 Glycoprotein X, putative n=1 Tax=Perkinsus mari... 57 1e-06
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 57 1e-06
UniRef100_B4PAZ8 DNA topoisomerase n=1 Tax=Drosophila yakuba Rep... 57 1e-06
UniRef100_B4L024 GI11736 n=1 Tax=Drosophila mojavensis RepID=B4L... 57 1e-06
UniRef100_B4JUL6 GH15522 n=1 Tax=Drosophila grimshawi RepID=B4JU... 57 1e-06
UniRef100_B4INW2 GM19323 n=1 Tax=Drosophila sechellia RepID=B4IN... 57 1e-06
UniRef100_B2VSJ8 Putative uncharacterized protein n=1 Tax=Pyreno... 57 1e-06
UniRef100_A8NE03 Predicted protein n=1 Tax=Coprinopsis cinerea o... 57 1e-06
UniRef100_B8GJ12 Periplasmic copper-binding n=1 Tax=Methanosphae... 57 1e-06
UniRef100_Q6IG02 Keratin, type II cytoskeletal 2 epidermal n=1 T... 57 1e-06
UniRef100_Q03173-5 Isoform 4 of Protein enabled homolog n=1 Tax=... 57 1e-06
UniRef100_Q03173-2 Isoform 1 of Protein enabled homolog n=1 Tax=... 57 1e-06
UniRef100_Q03173-3 Isoform 2 of Protein enabled homolog n=1 Tax=... 57 1e-06
UniRef100_Q03173-4 Isoform 3 of Protein enabled homolog n=1 Tax=... 57 1e-06
UniRef100_Q03173 Protein enabled homolog n=1 Tax=Mus musculus Re... 57 1e-06
UniRef100_UPI00017F00CB PREDICTED: similar to protocadherin 15 n... 56 1e-06
UniRef100_UPI000179D41E UPI000179D41E related cluster n=1 Tax=Bo... 56 1e-06
UniRef100_Q4RY48 Chromosome 3 SCAF14978, whole genome shotgun se... 56 1e-06
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 56 1e-06
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 56 1e-06
UniRef100_C1EVG8 Surface protein, pentapeptide repeat domain pro... 56 1e-06
UniRef100_B0TGZ6 Putative uncharacterized protein n=1 Tax=Heliob... 56 1e-06
UniRef100_Q9XER9 Putative transcription factor n=1 Tax=Arabidops... 56 1e-06
UniRef100_Q5VR46 Os01g0180000 protein n=1 Tax=Oryza sativa Japon... 56 1e-06
UniRef100_Q49I29 120 kDa pistil extensin-like protein (Fragment)... 56 1e-06
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 56 1e-06
UniRef100_C1N2F3 Predicted protein n=1 Tax=Micromonas pusilla CC... 56 1e-06
UniRef100_C1E755 Putative uncharacterized protein n=1 Tax=Microm... 56 1e-06
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_B8ADK4 Putative uncharacterized protein n=1 Tax=Oryza ... 56 1e-06
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 56 1e-06
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 56 1e-06
UniRef100_A7PES6 Chromosome chr11 scaffold_13, whole genome shot... 56 1e-06
UniRef100_B7Q3T8 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B7PVB4 Putative uncharacterized protein (Fragment) n=1... 56 1e-06
UniRef100_B4NCH1 GK25855 n=1 Tax=Drosophila willistoni RepID=B4N... 56 1e-06
UniRef100_B4L607 GI16284 n=1 Tax=Drosophila mojavensis RepID=B4L... 56 1e-06
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 56 1e-06
UniRef100_Q9Z0G8-2 Isoform 2 of WAS/WASL-interacting protein fam... 56 1e-06
UniRef100_Q9Z0G8 WAS/WASL-interacting protein family member 3 n=... 56 1e-06
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 56 1e-06
UniRef100_UPI0000F2BA2D PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 56 2e-06
UniRef100_UPI0000E20C65 PREDICTED: similar to Family with sequen... 56 2e-06
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 56 2e-06
UniRef100_O91332 EBNA-1 n=1 Tax=Macacine herpesvirus 4 RepID=O91... 56 2e-06
UniRef100_Q1BU84 Putative uncharacterized protein n=1 Tax=Burkho... 56 2e-06
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 56 2e-06
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 56 2e-06
UniRef100_A0K9V3 Putative uncharacterized protein n=1 Tax=Burkho... 56 2e-06
UniRef100_Q7Y215 Putative uncharacterized protein At2g05580 n=1 ... 56 2e-06
UniRef100_Q49I27 120 kDa pistil extensin-like protein (Fragment)... 56 2e-06
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 56 2e-06
UniRef100_A8HQ95 MATE efflux family protein n=1 Tax=Chlamydomona... 56 2e-06
UniRef100_A7QJD7 Chromosome chr8 scaffold_106, whole genome shot... 56 2e-06
UniRef100_A5AVJ0 Putative uncharacterized protein n=1 Tax=Vitis ... 56 2e-06
UniRef100_C3Z8C4 Putative uncharacterized protein n=1 Tax=Branch... 56 2e-06
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 56 2e-06
UniRef100_B6AF23 Putative uncharacterized protein n=1 Tax=Crypto... 56 2e-06
UniRef100_B4L4V7 GI14785 n=1 Tax=Drosophila mojavensis RepID=B4L... 56 2e-06
UniRef100_B4IVG9 GE14936 n=1 Tax=Drosophila yakuba RepID=B4IVG9_... 56 2e-06
UniRef100_B4IN23 GM19303 n=1 Tax=Drosophila sechellia RepID=B4IN... 56 2e-06
UniRef100_B4IMQ0 GM13143 n=1 Tax=Drosophila sechellia RepID=B4IM... 56 2e-06
UniRef100_C9J285 Putative uncharacterized protein ENSP0000039240... 56 2e-06
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 56 2e-06
UniRef100_B6QV40 Cytokinesis protein SepA/Bni1 n=1 Tax=Penicilli... 56 2e-06
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI000186D2E7 hypothetical protein Phum_PHUM146270 n=1... 55 2e-06
UniRef100_UPI000155BA60 PREDICTED: similar to hCG2029577, partia... 55 2e-06
UniRef100_UPI0000DD90BE Os04g0438100 n=1 Tax=Oryza sativa Japoni... 55 2e-06
UniRef100_UPI00005A34B4 PREDICTED: hypothetical protein XP_85934... 55 2e-06
UniRef100_Q4A373 Putative lectin protein n=2 Tax=Emiliania huxle... 55 2e-06
UniRef100_A9VSK7 LPXTG-motif cell wall anchor domain n=1 Tax=Bac... 55 2e-06
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 55 2e-06
UniRef100_A1B9K3 FHA domain containing protein n=1 Tax=Paracoccu... 55 2e-06
UniRef100_Q9FPQ5 Gamete-specific hydroxyproline-rich glycoprotei... 55 2e-06
UniRef100_Q4PSF3 Proline-rich extensin-like family protein n=1 T... 55 2e-06
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 55 2e-06
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 55 2e-06
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 55 2e-06
UniRef100_B9P7M7 Predicted protein n=1 Tax=Populus trichocarpa R... 55 2e-06
UniRef100_A8HY82 Predicted protein n=1 Tax=Chlamydomonas reinhar... 55 2e-06
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 55 2e-06
UniRef100_Q5CFZ6 Putative uncharacterized protein n=1 Tax=Crypto... 55 2e-06
UniRef100_Q4DD51 Putative uncharacterized protein n=1 Tax=Trypan... 55 2e-06
UniRef100_C4QFT9 Diaphanous, putative n=1 Tax=Schistosoma manson... 55 2e-06
UniRef100_B7PQ78 H/ACA ribonucleoprotein complex protein, putati... 55 2e-06
UniRef100_B4MTQ5 GK23886 n=1 Tax=Drosophila willistoni RepID=B4M... 55 2e-06
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 55 2e-06
UniRef100_Q6C5H5 YALI0E18007p n=1 Tax=Yarrowia lipolytica RepID=... 55 2e-06
UniRef100_C1H633 Predicted protein n=1 Tax=Paracoccidioides bras... 55 2e-06
UniRef100_A4QSC9 Predicted protein n=1 Tax=Magnaporthe grisea Re... 55 2e-06
UniRef100_A8M746 Translation initiation factor IF-2 n=1 Tax=Sali... 55 2e-06
UniRef100_UPI000186A01D hypothetical protein BRAFLDRAFT_106142 n... 55 3e-06
UniRef100_UPI0000F2E115 PREDICTED: similar to Dachshund homolog ... 55 3e-06
UniRef100_UPI0000F2D0CF PREDICTED: similar to reverse transcript... 55 3e-06
UniRef100_UPI0000DA1EE3 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0000DA1CBA PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 55 3e-06
UniRef100_UPI000179D3E7 UPI000179D3E7 related cluster n=1 Tax=Bo... 55 3e-06
UniRef100_Q06KR7 Viral capsid associated protein n=1 Tax=Anticar... 55 3e-06
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 55 3e-06
UniRef100_B7H7J6 Surface protein n=1 Tax=Bacillus cereus B4264 R... 55 3e-06
UniRef100_B3ZQ61 Lpxtg-motif cell wall anchor domain protein n=1... 55 3e-06
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 55 3e-06
UniRef100_Q5ZB68 Os01g0594300 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 3e-06
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 3e-06
UniRef100_Q55F61 Putative uncharacterized protein n=1 Tax=Dictyo... 55 3e-06
UniRef100_Q3HVK1 Hoglet n=1 Tax=Monosiga ovata RepID=Q3HVK1_9EUKA 55 3e-06
UniRef100_C5K9U4 Putative uncharacterized protein n=1 Tax=Perkin... 55 3e-06
UniRef100_C0MP97 Formin-homology protein SmDia n=1 Tax=Schistoso... 55 3e-06
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 55 3e-06
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 55 3e-06
UniRef100_B7P0W1 Cement protein, putative (Fragment) n=1 Tax=Ixo... 55 3e-06
UniRef100_B4NPA0 GK17577 n=1 Tax=Drosophila willistoni RepID=B4N... 55 3e-06
UniRef100_B4NLW8 GK10515 n=1 Tax=Drosophila willistoni RepID=B4N... 55 3e-06
UniRef100_B4MT24 GK19787 n=1 Tax=Drosophila willistoni RepID=B4M... 55 3e-06
UniRef100_B4KDH9 GI22470 n=1 Tax=Drosophila mojavensis RepID=B4K... 55 3e-06
UniRef100_B4IT39 GE18257 n=1 Tax=Drosophila yakuba RepID=B4IT39_... 55 3e-06
UniRef100_A9VE65 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 3e-06
UniRef100_A9V6K4 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 3e-06
UniRef100_Q2H4F1 Putative uncharacterized protein n=1 Tax=Chaeto... 55 3e-06
UniRef100_B6K7J0 Predicted protein n=1 Tax=Schizosaccharomyces j... 55 3e-06
UniRef100_B0XR50 Serine-threonine rich protein, putative n=1 Tax... 55 3e-06
UniRef100_A2R0A5 Remark: C-terminal truncated ORF due to end of ... 55 3e-06
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0001863C23 hypothetical protein BRAFLDRAFT_82685 n=... 55 4e-06
UniRef100_UPI0001795912 PREDICTED: similar to protocadherin 15 n... 55 4e-06
UniRef100_UPI0000E48096 PREDICTED: hypothetical protein n=1 Tax=... 55 4e-06
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_UPI0000E12297 Os03g0813200 n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_UPI0000E1203E Os03g0308700 n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_UPI00004A413C PREDICTED: similar to WAS protein family... 55 4e-06
UniRef100_Q4SVG8 Chromosome undetermined SCAF13758, whole genome... 55 4e-06
UniRef100_Q8V0M0 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 55 4e-06
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 55 4e-06
UniRef100_Q2K740 Hypothetical conserved protein n=1 Tax=Rhizobiu... 55 4e-06
UniRef100_C2RHX6 Putative uncharacterized protein n=1 Tax=Bacill... 55 4e-06
UniRef100_B3ZAP2 Lpxtg-motif cell wall anchor domain protein n=1... 55 4e-06
UniRef100_A3IUY3 Putative uncharacterized protein n=1 Tax=Cyanot... 55 4e-06
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 55 4e-06
UniRef100_Q75HG8 Putative prohibitin n=1 Tax=Oryza sativa Japoni... 55 4e-06
UniRef100_Q4U2V9 Hydroxyproline-rich glycoprotein GAS30 n=1 Tax=... 55 4e-06
UniRef100_Q10R38 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_Q0DME4 Os03g0813200 protein n=1 Tax=Oryza sativa Japon... 55 4e-06
UniRef100_B9GW18 Predicted protein n=1 Tax=Populus trichocarpa R... 55 4e-06
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 55 4e-06
UniRef100_A9SB49 Predicted protein n=1 Tax=Physcomitrella patens... 55 4e-06
UniRef100_Q6UDW6 Erythrocyte membrane protein 1 n=1 Tax=Plasmodi... 55 4e-06
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 55 4e-06
UniRef100_B4LU87 GJ17267 n=1 Tax=Drosophila virilis RepID=B4LU87... 55 4e-06
UniRef100_B3N5G6 GG25361, isoform A n=1 Tax=Drosophila erecta Re... 55 4e-06
UniRef100_A8Y2E4 Putative uncharacterized protein (Fragment) n=1... 55 4e-06
UniRef100_A7UN18 Acp27b (Fragment) n=1 Tax=Drosophila mojavensis... 55 4e-06
UniRef100_A5JUU8 Formin B n=2 Tax=Trypanosoma brucei RepID=A5JUU... 55 4e-06
UniRef100_Q4WC06 Serine-threonine rich protein, putative n=1 Tax... 55 4e-06
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 55 4e-06
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 55 4e-06
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 55 4e-06
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 55 4e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 55 4e-06
UniRef100_UPI0001924FB1 PREDICTED: hypothetical protein, partial... 54 5e-06
UniRef100_UPI000180B79E PREDICTED: similar to Ras association an... 54 5e-06
UniRef100_UPI000155C73A PREDICTED: similar to proline rich 8 n=1... 54 5e-06
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI0000DA2F5F PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI0000DA1EB9 PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI0000D9ED26 PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI00004296C7 SET binding protein 1 n=2 Tax=Mus muscul... 54 5e-06
UniRef100_UPI00015DF6E5 enabled homolog (Drosophila) n=1 Tax=Mus... 54 5e-06
UniRef100_UPI00015DF9DB Proline-rich protein 12. n=1 Tax=Homo sa... 54 5e-06
UniRef100_UPI0001596889 proline rich 12 n=1 Tax=Homo sapiens Rep... 54 5e-06
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 54 5e-06
UniRef100_Q4SUB2 Chromosome 3 SCAF13974, whole genome shotgun se... 54 5e-06
UniRef100_Q8V0L5 Glycoprotein gp2 n=1 Tax=Equid herpesvirus 1 Re... 54 5e-06
UniRef100_O39782 Membrane glycoprotein n=1 Tax=Equid herpesvirus... 54 5e-06
UniRef100_O39781 Membrane glycoprotein n=1 Tax=Equid herpesvirus... 54 5e-06
UniRef100_B1WBQ8 Glyceraldehyde 3-phosphate dehydrogenase n=1 Ta... 54 5e-06
UniRef100_Q6HNZ6 Surface protein, pentapeptide repeat domain n=1... 54 5e-06
UniRef100_Q2JMC8 TonB family protein n=1 Tax=Synechococcus sp. J... 54 5e-06
UniRef100_B2HP21 Conserved hypothetical membrane protein n=1 Tax... 54 5e-06
UniRef100_A7INX6 Putative head morphogenesis protein SPP1 gp7 n=... 54 5e-06
UniRef100_A0R9A1 Surface protein, pentapeptide repeat domain n=1... 54 5e-06
UniRef100_Q2QR13 Retrotransposon protein, putative, Ty3-gypsy su... 54 5e-06
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 54 5e-06
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 54 5e-06
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 54 5e-06
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_A8IRQ5 DEAD/DEAH box helicase n=1 Tax=Chlamydomonas re... 54 5e-06
UniRef100_A3CH91 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_A7Z068 LMOD2 protein n=1 Tax=Bos taurus RepID=A7Z068_B... 54 5e-06
UniRef100_Q7YYP6 Hydroxyproline-rich glycoprotein dz-hrgp, proba... 54 5e-06
UniRef100_Q68KY8 Optomotor blind (Fragment) n=1 Tax=Drosophila p... 54 5e-06
UniRef100_Q38EF1 Putative uncharacterized protein n=1 Tax=Trypan... 54 5e-06
UniRef100_Q18444 Putative uncharacterized protein n=1 Tax=Caenor... 54 5e-06
UniRef100_O15647 Fibrillarin (Fragment) n=1 Tax=Plasmodium falci... 54 5e-06
UniRef100_C9ZY65 Putative uncharacterized protein n=1 Tax=Trypan... 54 5e-06
UniRef100_C5LG38 Merozoite surface protein 2, putative n=1 Tax=P... 54 5e-06
UniRef100_C5LA81 Putative uncharacterized protein n=1 Tax=Perkin... 54 5e-06
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 54 5e-06
UniRef100_B4MDK8 GJ16154 n=1 Tax=Drosophila virilis RepID=B4MDK8... 54 5e-06
UniRef100_B4KPH5 GI18675 n=1 Tax=Drosophila mojavensis RepID=B4K... 54 5e-06
UniRef100_B4IJN4 GM17713 (Fragment) n=1 Tax=Drosophila sechellia... 54 5e-06
UniRef100_A8Q0Y3 CG12586-PA, putative n=1 Tax=Brugia malayi RepI... 54 5e-06
UniRef100_A8PT37 Cell wall protein DAN4, putative n=1 Tax=Brugia... 54 5e-06
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 54 5e-06
UniRef100_A6NFN6 Putative uncharacterized protein PRR12 n=1 Tax=... 54 5e-06
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 54 5e-06
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 54 5e-06
UniRef100_C0NKI4 Basic proline-rich protein n=1 Tax=Ajellomyces ... 54 5e-06
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 54 5e-06
UniRef100_A6QWU5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 54 5e-06
UniRef100_B7R1V6 Metallophosphoesterase, calcineurin superfamily... 54 5e-06
UniRef100_Q9Z180 SET-binding protein n=1 Tax=Mus musculus RepID=... 54 5e-06
UniRef100_Q9ULL5-2 Isoform 2 of Proline-rich protein 12 n=1 Tax=... 54 5e-06
UniRef100_Q9ULL5 Proline-rich protein 12 n=1 Tax=Homo sapiens Re... 54 5e-06
UniRef100_Q6S6W0 Glycoprotein gp2 n=1 Tax=Equine herpesvirus 1 (... 54 5e-06
UniRef100_P28968 Glycoprotein gp2 n=1 Tax=Equine herpesvirus typ... 54 5e-06
UniRef100_Q9ESV6 Glyceraldehyde-3-phosphate dehydrogenase, testi... 54 5e-06
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 54 5e-06
UniRef100_UPI000194C58F PREDICTED: formin 1 n=1 Tax=Taeniopygia ... 54 6e-06
UniRef100_UPI000194C22E PREDICTED: similar to formin 2 n=1 Tax=T... 54 6e-06
UniRef100_UPI0001925DB5 PREDICTED: similar to Wiskott-Aldrich sy... 54 6e-06
UniRef100_UPI0001924E84 PREDICTED: similar to Wiskott-Aldrich sy... 54 6e-06
UniRef100_UPI000180D259 PREDICTED: similar to WAS protein family... 54 6e-06
UniRef100_UPI00016B0970 putative polyketide synthase n=1 Tax=Bur... 54 6e-06
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000F2E9FE PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000F2C8C4 PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000DA3D7B PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000DA36CE PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000DA1FBD PREDICTED: hypothetical protein n=1 Tax=... 54 6e-06
UniRef100_UPI0000D9CC91 PREDICTED: similar to keratin 1 isoform ... 54 6e-06
UniRef100_UPI0000D9B773 PREDICTED: similar to CG5514-PB, isoform... 54 6e-06
UniRef100_Q69ZN8 MKIAA1205 protein (Fragment) n=3 Tax=Mus muscul... 54 6e-06
UniRef100_UPI00017B5A37 UPI00017B5A37 related cluster n=1 Tax=Te... 54 6e-06
UniRef100_UPI00017B5A36 UPI00017B5A36 related cluster n=1 Tax=Te... 54 6e-06
UniRef100_UPI00017B5A35 UPI00017B5A35 related cluster n=1 Tax=Te... 54 6e-06
UniRef100_UPI000060414B formin-like domain containing protein MA... 54 6e-06
UniRef100_UPI00016E37F3 UPI00016E37F3 related cluster n=1 Tax=Ta... 54 6e-06
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 54 6e-06
UniRef100_Q1AMQ0 Antifreeze glycoprotein n=1 Tax=Eleginus gracil... 54 6e-06
UniRef100_O13028 Antifreeze glycopeptide AFGP polyprotein n=1 Ta... 54 6e-06
UniRef100_B1H2Z4 LOC100145517 protein n=1 Tax=Xenopus (Silurana)... 54 6e-06
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 54 6e-06
UniRef100_B7FED2 Envelope glycoprotein n=1 Tax=Equid herpesvirus... 54 6e-06
UniRef100_B2RTP7 Krt2 protein n=1 Tax=Mus musculus RepID=B2RTP7_... 54 6e-06
UniRef100_B1JXE0 Putative uncharacterized protein n=1 Tax=Burkho... 54 6e-06
UniRef100_C7QJM7 RNA binding S1 domain protein n=1 Tax=Catenulis... 54 6e-06
UniRef100_B4D3Z5 Putative uncharacterized protein n=1 Tax=Chthon... 54 6e-06
UniRef100_A0YXE4 Putative uncharacterized protein n=1 Tax=Lyngby... 54 6e-06
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 54 6e-06
UniRef100_Q948Y9 VMP1 protein n=1 Tax=Volvox carteri f. nagarien... 54 6e-06
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 54 6e-06
UniRef100_Q49I31 120 kDa pistil extensin-like protein (Fragment)... 54 6e-06
UniRef100_Q41188 Glycine-rich protein n=1 Tax=Arabidopsis thalia... 54 6e-06
UniRef100_C5XQF9 Putative uncharacterized protein Sb03g040290 n=... 54 6e-06
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 54 6e-06
UniRef100_B9HJA5 Predicted protein n=1 Tax=Populus trichocarpa R... 54 6e-06
UniRef100_B9H7Q5 Predicted protein n=1 Tax=Populus trichocarpa R... 54 6e-06
UniRef100_Q2QLY3 Os12g0624100 protein n=2 Tax=Oryza sativa RepID... 54 6e-06
UniRef100_A9RI93 Predicted protein n=1 Tax=Physcomitrella patens... 54 6e-06
UniRef100_Q9TZM9 Temporarily assigned gene name protein 343, iso... 54 6e-06
UniRef100_Q675X6 Putative uncharacterized protein n=1 Tax=Oikopl... 54 6e-06
UniRef100_Q5CW96 Signal peptide plus His and thr repeats, signal... 54 6e-06
UniRef100_Q5CRW5 Putative uncharacterized protein n=1 Tax=Crypto... 54 6e-06
UniRef100_Q1XFY2 Temporarily assigned gene name protein 343, iso... 54 6e-06
UniRef100_Q16TF1 Fibrillarin (Fragment) n=1 Tax=Aedes aegypti Re... 54 6e-06
UniRef100_B7QDE3 Putative uncharacterized protein (Fragment) n=1... 54 6e-06
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 54 6e-06
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 54 6e-06
UniRef100_B4JSS1 GH22875 n=1 Tax=Drosophila grimshawi RepID=B4JS... 54 6e-06
UniRef100_B4JSH2 GH18083 n=1 Tax=Drosophila grimshawi RepID=B4JS... 54 6e-06
UniRef100_B4J5L7 GH21076 n=1 Tax=Drosophila grimshawi RepID=B4J5... 54 6e-06
UniRef100_B3MRZ2 GF20902 n=1 Tax=Drosophila ananassae RepID=B3MR... 54 6e-06
UniRef100_B3MAN6 GF10963 n=1 Tax=Drosophila ananassae RepID=B3MA... 54 6e-06
UniRef100_A9V3F3 Predicted protein n=1 Tax=Monosiga brevicollis ... 54 6e-06
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 54 6e-06
UniRef100_B4E3J5 cDNA FLJ55445, highly similar to Wiskott-Aldric... 54 6e-06
UniRef100_C6HGV1 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 54 6e-06
UniRef100_C0NZQ8 Cytokinesis protein SepA/Bni1 n=1 Tax=Ajellomyc... 54 6e-06
UniRef100_A1CLV3 Putative uncharacterized protein n=1 Tax=Asperg... 54 6e-06
UniRef100_A3MV74 Putative uncharacterized protein n=1 Tax=Pyroba... 54 6e-06
UniRef100_Q9Y6W5 Wiskott-Aldrich syndrome protein family member ... 54 6e-06
UniRef100_A2APV2-3 Isoform 3 of Formin-like protein 2 n=1 Tax=Mu... 54 6e-06
UniRef100_A2APV2 Formin-like protein 2 n=1 Tax=Mus musculus RepI... 54 6e-06
UniRef100_UPI0001985275 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI00017923D4 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI00017466C4 RNA-binding protein (RRM domain) n=1 Tax... 54 8e-06
UniRef100_UPI00015B5EB5 PREDICTED: similar to CG3606-PB n=1 Tax=... 54 8e-06
UniRef100_UPI00015539C6 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI00015539BB PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000E47C3A PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000E468D6 PREDICTED: similar to laminin alpha chai... 54 8e-06
UniRef100_UPI0000DB7FFD PREDICTED: similar to CG18497-PA, isofor... 54 8e-06
UniRef100_UPI0000DA4604 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000DA452D PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000DA421E PREDICTED: similar to SET binding protei... 54 8e-06
UniRef100_UPI0000DA2519 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000DA20D4 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI0000DA1F29 PREDICTED: hypothetical protein n=1 Tax=... 54 8e-06
UniRef100_UPI000069F77D Mucin n=1 Tax=Xenopus (Silurana) tropica... 54 8e-06
UniRef100_UPI0001B7A6BD enabled homolog n=1 Tax=Rattus norvegicu... 54 8e-06
UniRef100_UPI0001B7A6BC enabled homolog n=1 Tax=Rattus norvegicu... 54 8e-06
UniRef100_UPI0001B7A6A7 enabled homolog n=1 Tax=Rattus norvegicu... 54 8e-06
UniRef100_Q9Y6X0 SET-binding protein n=2 Tax=Homo sapiens RepID=... 54 8e-06
UniRef100_UPI00016E4781 UPI00016E4781 related cluster n=1 Tax=Ta... 54 8e-06
UniRef100_Q8V0M2 Glycoprotein gp2 (Fragment) n=1 Tax=Equid herpe... 54 8e-06
UniRef100_Q5XHX3 Enabled homolog (Drosophila) n=1 Tax=Rattus nor... 54 8e-06
UniRef100_Q0RKP1 Putative Chaperone protein dnaK (Heat shock pro... 54 8e-06