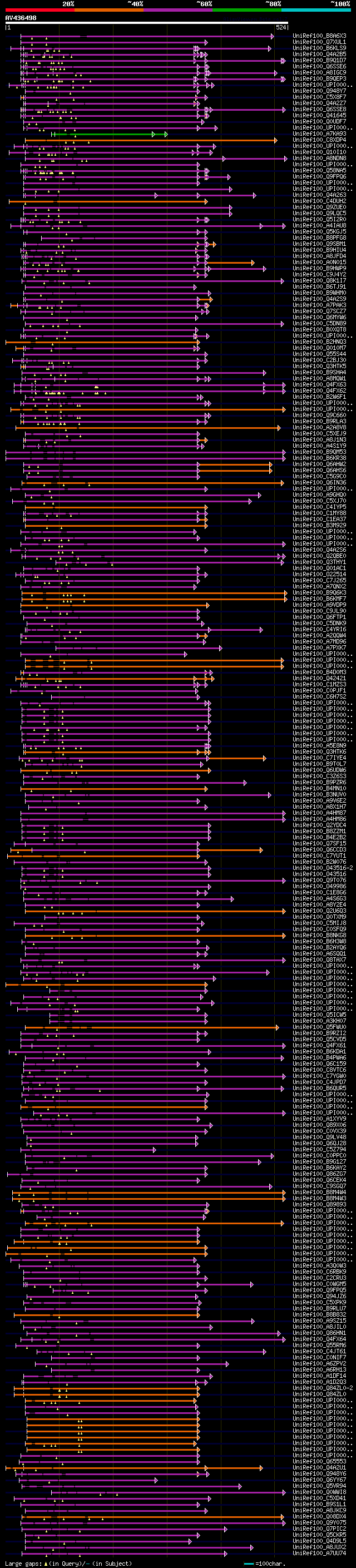
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV436498 PS003c09_r
(524 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B8A6X3 Putative uncharacterized protein n=1 Tax=Oryza ... 78 4e-13
UniRef100_Q7XUL1 Os04g0498700 protein n=1 Tax=Oryza sativa Japon... 70 9e-11
UniRef100_B6KLS9 Voltage gated chloride channel domain-containin... 69 2e-10
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 68 4e-10
UniRef100_B9Q1D7 Chloride channel, putative n=1 Tax=Toxoplasma g... 68 4e-10
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 67 7e-10
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 66 2e-09
UniRef100_B9QEP3 Chloride channel protein k, putative n=1 Tax=To... 66 2e-09
UniRef100_UPI0001A7B3C7 unknown protein n=1 Tax=Arabidopsis thal... 65 3e-09
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 65 3e-09
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 64 5e-09
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 64 6e-09
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 64 6e-09
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 64 8e-09
UniRef100_Q0UDF7 Putative uncharacterized protein n=1 Tax=Phaeos... 64 8e-09
UniRef100_UPI00006A1DCA UPI00006A1DCA related cluster n=1 Tax=Xe... 63 1e-08
UniRef100_A7KA93 Putative uncharacterized protein Z833L n=1 Tax=... 63 1e-08
UniRef100_C8XDP4 Serine/threonine protein kinase n=1 Tax=Nakamur... 63 1e-08
UniRef100_UPI00006A1DCC UPI00006A1DCC related cluster n=1 Tax=Xe... 63 1e-08
UniRef100_Q10I10 Os03g0568800 protein n=1 Tax=Oryza sativa Japon... 63 1e-08
UniRef100_A8NDN8 Predicted protein n=1 Tax=Coprinopsis cinerea o... 63 1e-08
UniRef100_UPI0000E25E91 PREDICTED: similar to WD repeat domain 4... 62 2e-08
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 62 2e-08
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 62 2e-08
UniRef100_UPI0001868875 hypothetical protein BRAFLDRAFT_103538 n... 62 2e-08
UniRef100_UPI000034F3D3 protein kinase family protein n=1 Tax=Ar... 62 2e-08
UniRef100_Q4A263 Putative membrane protein n=1 Tax=Emiliania hux... 62 2e-08
UniRef100_C4DUH2 Putative uncharacterized protein n=1 Tax=Stacke... 62 2e-08
UniRef100_Q9ZUE0 F5O8.10 protein n=1 Tax=Arabidopsis thaliana Re... 62 2e-08
UniRef100_Q9LQC5 F28C11.17 n=1 Tax=Arabidopsis thaliana RepID=Q9... 62 2e-08
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 62 3e-08
UniRef100_A4IAU8 Proteophosphoglycan ppg4 n=1 Tax=Leishmania inf... 62 3e-08
UniRef100_Q5KGJ5 Putative uncharacterized protein n=1 Tax=Filoba... 62 3e-08
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 62 3e-08
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 61 4e-08
UniRef100_B9HIU4 Predicted protein (Fragment) n=1 Tax=Populus tr... 61 4e-08
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 61 4e-08
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 61 4e-08
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 61 5e-08
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 61 5e-08
UniRef100_Q8K1I7 WAS/WASL-interacting protein family member 1 n=... 61 5e-08
UniRef100_B6TJ91 Splicing factor, arginine/serine-rich 7 n=1 Tax... 60 7e-08
UniRef100_B9WHM0 Vegetative cell wall protein, putative (Hydroxy... 60 7e-08
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 60 9e-08
UniRef100_A7PAK3 Chromosome chr14 scaffold_9, whole genome shotg... 60 9e-08
UniRef100_Q7SCZ7 WASP-interacting protein-like protein vrp1p n=1... 60 9e-08
UniRef100_Q6MYW6 Basic proline-rich protein n=1 Tax=Aspergillus ... 60 9e-08
UniRef100_C5DN89 KLTH0G15048p n=1 Tax=Lachancea thermotolerans C... 60 9e-08
UniRef100_B0XQT8 Conserved proline-rich protein n=2 Tax=Aspergil... 60 9e-08
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_B2HNQ3 Conserved hypothetical alanine and proline rich... 60 1e-07
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 60 1e-07
UniRef100_Q55S44 Putative uncharacterized protein n=1 Tax=Filoba... 60 1e-07
UniRef100_C2BJ30 Fe-S oxidoreductase n=1 Tax=Corynebacterium pse... 59 2e-07
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 59 2e-07
UniRef100_B9SHA4 ATP binding protein, putative n=1 Tax=Ricinus c... 59 2e-07
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 59 2e-07
UniRef100_Q4FX63 Proteophosphoglycan ppg4 n=1 Tax=Leishmania maj... 59 2e-07
UniRef100_Q4FX62 Proteophosphoglycan 5 n=1 Tax=Leishmania major ... 59 2e-07
UniRef100_B2W6F1 Putative uncharacterized protein n=1 Tax=Pyreno... 59 2e-07
UniRef100_UPI00005DC1BB PERK10 (PROLINE-RICH EXTENSIN-LIKE RECEP... 59 2e-07
UniRef100_UPI0000DC00C7 UPI0000DC00C7 related cluster n=1 Tax=Ra... 59 2e-07
UniRef100_Q9C660 Pto kinase interactor, putative n=1 Tax=Arabido... 59 2e-07
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 59 2e-07
UniRef100_A2A8V8 Serine/arginine repetitive matrix 1 n=1 Tax=Mus... 59 3e-07
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 59 3e-07
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 59 3e-07
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 59 3e-07
UniRef100_B9QM53 Sodium/hydrogen exchanger n=1 Tax=Toxoplasma go... 59 3e-07
UniRef100_B6KR38 Sodium/hydrogen exchanger n=1 Tax=Toxoplasma go... 59 3e-07
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 59 3e-07
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 59 3e-07
UniRef100_C5G9C0 Putative uncharacterized protein n=1 Tax=Ajello... 59 3e-07
UniRef100_Q6IN36 WAS/WASL-interacting protein family member 1 n=... 59 3e-07
UniRef100_UPI00017B2A82 UPI00017B2A82 related cluster n=1 Tax=Te... 58 3e-07
UniRef100_A9GHQ0 Cellulose synthase 1 operon protein C n=1 Tax=S... 58 3e-07
UniRef100_C5XJ70 Putative uncharacterized protein Sb03g034040 n=... 58 3e-07
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 58 3e-07
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 58 3e-07
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 58 3e-07
UniRef100_B3M929 GF25073 n=1 Tax=Drosophila ananassae RepID=B3M9... 58 3e-07
UniRef100_UPI00019829F7 PREDICTED: hypothetical protein n=1 Tax=... 58 5e-07
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 58 5e-07
UniRef100_UPI000013E9DD mucin 7, secreted precursor n=1 Tax=Homo... 58 5e-07
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 58 5e-07
UniRef100_Q2QBE0 UL36 n=1 Tax=Papiine herpesvirus 2 RepID=Q2QBE0... 58 5e-07
UniRef100_Q3THY1 Putative uncharacterized protein n=1 Tax=Mus mu... 58 5e-07
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 58 5e-07
UniRef100_O22514 Proline rich protein n=1 Tax=Santalum album Rep... 58 5e-07
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 58 5e-07
UniRef100_A7QNX2 Chromosome chr1 scaffold_135, whole genome shot... 58 5e-07
UniRef100_B9Q6K3 Putative uncharacterized protein n=1 Tax=Toxopl... 58 5e-07
UniRef100_B6KMF7 Putative uncharacterized protein n=1 Tax=Toxopl... 58 5e-07
UniRef100_A9VDP9 Predicted protein n=1 Tax=Monosiga brevicollis ... 58 5e-07
UniRef100_C9JL90 Putative uncharacterized protein WIPF1 n=1 Tax=... 58 5e-07
UniRef100_Q6FTP1 Similar to uniprot|P37370 Saccharomyces cerevis... 58 5e-07
UniRef100_C5DNK9 KLTH0G17886p n=1 Tax=Lachancea thermotolerans C... 58 5e-07
UniRef100_C4YRI6 Predicted protein n=1 Tax=Candida albicans RepI... 58 5e-07
UniRef100_A2QQW4 Contig An08c0110, complete genome n=1 Tax=Asper... 58 5e-07
UniRef100_A7MD96 SYNPO protein (Fragment) n=2 Tax=Homo sapiens R... 58 5e-07
UniRef100_A7PXK7 Chromosome chr12 scaffold_36, whole genome shot... 57 6e-07
UniRef100_UPI0000E121D9 Os03g0717300 n=1 Tax=Oryza sativa Japoni... 57 6e-07
UniRef100_UPI00005A5941 PREDICTED: similar to Wiskott-Aldrich sy... 57 6e-07
UniRef100_UPI0000EB01B2 WAS/WASL interacting protein family memb... 57 6e-07
UniRef100_B4D0M3 Putative uncharacterized protein n=1 Tax=Chthon... 57 6e-07
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 57 6e-07
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 57 6e-07
UniRef100_C0PJF1 Putative uncharacterized protein n=1 Tax=Zea ma... 57 6e-07
UniRef100_C6H7S2 Putative uncharacterized protein n=1 Tax=Ajello... 57 6e-07
UniRef100_UPI0001B55E13 putative chaplin n=1 Tax=Streptomyces sp... 57 8e-07
UniRef100_UPI0000E1F801 PREDICTED: similar to WASPIP protein iso... 57 8e-07
UniRef100_UPI0000E1F800 PREDICTED: similar to SH3-domain interac... 57 8e-07
UniRef100_UPI0000E1F7FD PREDICTED: similar to WASP interacting p... 57 8e-07
UniRef100_UPI0000DB7674 PREDICTED: hypothetical protein n=1 Tax=... 57 8e-07
UniRef100_UPI0001881868 UPI0001881868 related cluster n=1 Tax=Ho... 57 8e-07
UniRef100_UPI0000D61270 WAS/WASL interacting protein family memb... 57 8e-07
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 57 8e-07
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 57 8e-07
UniRef100_C7IYE4 Os02g0557000 protein (Fragment) n=1 Tax=Oryza s... 57 8e-07
UniRef100_B9T0L7 Nutrient reservoir, putative n=1 Tax=Ricinus co... 57 8e-07
UniRef100_Q6UDW6 Erythrocyte membrane protein 1 n=1 Tax=Plasmodi... 57 8e-07
UniRef100_C3Z6S3 Putative uncharacterized protein n=1 Tax=Branch... 57 8e-07
UniRef100_B9PZR6 Putative uncharacterized protein n=1 Tax=Toxopl... 57 8e-07
UniRef100_B4MN10 GK16584 n=1 Tax=Drosophila willistoni RepID=B4M... 57 8e-07
UniRef100_B3NUV0 GG17864 n=1 Tax=Drosophila erecta RepID=B3NUV0_... 57 8e-07
UniRef100_A9V6E2 Predicted protein n=1 Tax=Monosiga brevicollis ... 57 8e-07
UniRef100_A8X1H7 Putative uncharacterized protein n=1 Tax=Caenor... 57 8e-07
UniRef100_A4HM87 Proteophosphoglycan ppg4 n=1 Tax=Leishmania bra... 57 8e-07
UniRef100_A4HM86 Proteophosphoglycan ppg4 n=1 Tax=Leishmania bra... 57 8e-07
UniRef100_Q2YDC4 WIPF1 protein n=1 Tax=Homo sapiens RepID=Q2YDC4... 57 8e-07
UniRef100_B8ZZM1 Putative uncharacterized protein WIPF1 n=1 Tax=... 57 8e-07
UniRef100_B4E2B2 cDNA FLJ61061, highly similar to Wiskott-Aldric... 57 8e-07
UniRef100_Q7SF15 WASP-like pretein las17p n=1 Tax=Neurospora cra... 57 8e-07
UniRef100_Q6CCD3 YALI0C10450p n=1 Tax=Yarrowia lipolytica RepID=... 57 8e-07
UniRef100_C7YUT1 Putative uncharacterized protein n=1 Tax=Nectri... 57 8e-07
UniRef100_B2W076 Predicted protein n=1 Tax=Pyrenophora tritici-r... 57 8e-07
UniRef100_O43516-2 Isoform 2 of WAS/WASL-interacting protein fam... 57 8e-07
UniRef100_O43516 WAS/WASL-interacting protein family member 1 n=... 57 8e-07
UniRef100_Q9T076 Early nodulin-like protein 2 n=1 Tax=Arabidopsi... 57 8e-07
UniRef100_O49986 120 kDa style glycoprotein n=1 Tax=Nicotiana al... 57 1e-06
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 57 1e-06
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 57 1e-06
UniRef100_A8Y2E4 Putative uncharacterized protein (Fragment) n=1... 57 1e-06
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 57 1e-06
UniRef100_Q0TXM9 Putative uncharacterized protein n=1 Tax=Phaeos... 57 1e-06
UniRef100_C5MIJ8 Predicted protein n=1 Tax=Candida tropicalis MY... 57 1e-06
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 57 1e-06
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 57 1e-06
UniRef100_B6H3W8 Pc13g11020 protein n=1 Tax=Penicillium chrysoge... 57 1e-06
UniRef100_B2AYQ6 Predicted CDS Pa_1_11890 n=1 Tax=Podospora anse... 57 1e-06
UniRef100_A6SQQ1 Putative uncharacterized protein n=1 Tax=Botryo... 57 1e-06
UniRef100_Q8TAX7 Mucin-7 n=1 Tax=Homo sapiens RepID=MUC7_HUMAN 57 1e-06
UniRef100_UPI0001925155 PREDICTED: hypothetical protein, partial... 56 1e-06
UniRef100_UPI00017F03E3 PREDICTED: similar to Ras and Rab intera... 56 1e-06
UniRef100_UPI0000E240CF PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI0000D9360A PREDICTED: similar to SH3 domain-binding... 56 1e-06
UniRef100_UPI0001A2C549 protocadherin 15b n=1 Tax=Danio rerio Re... 56 1e-06
UniRef100_UPI000069F8DC UPI000069F8DC related cluster n=1 Tax=Xe... 56 1e-06
UniRef100_UPI00017B2A80 UPI00017B2A80 related cluster n=1 Tax=Te... 56 1e-06
UniRef100_UPI000162C860 nascent polypeptide-associated complex a... 56 1e-06
UniRef100_Q5ICW5 Protocadherin 15b n=1 Tax=Danio rerio RepID=Q5I... 56 1e-06
UniRef100_A3KH07 Protocadherin 15b (Fragment) n=1 Tax=Danio reri... 56 1e-06
UniRef100_Q5FWU0 WAS protein family, member 2 n=1 Tax=Rattus nor... 56 1e-06
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 56 1e-06
UniRef100_Q5CVD5 Putative uncharacterized protein n=1 Tax=Crypto... 56 1e-06
UniRef100_Q4FX61 Proteophosphoglycan ppg1 n=2 Tax=Leishmania maj... 56 1e-06
UniRef100_B6KDA1 Putative uncharacterized protein n=1 Tax=Toxopl... 56 1e-06
UniRef100_B4PWA6 GE17167 n=1 Tax=Drosophila yakuba RepID=B4PWA6_... 56 1e-06
UniRef100_Q6C159 YALI0F19030p n=1 Tax=Yarrowia lipolytica RepID=... 56 1e-06
UniRef100_C8VTC6 Proline-rich, actin-associated protein Vrp1, pu... 56 1e-06
UniRef100_C7YGW0 Putative uncharacterized protein n=1 Tax=Nectri... 56 1e-06
UniRef100_C4JPD7 Putative uncharacterized protein n=1 Tax=Uncino... 56 1e-06
UniRef100_B6QUR5 Putative uncharacterized protein n=1 Tax=Penici... 56 1e-06
UniRef100_UPI000194AF17 protein DR1 n=1 Tax=Human herpesvirus 6 ... 56 2e-06
UniRef100_UPI0001925A99 PREDICTED: hypothetical protein, partial... 56 2e-06
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 56 2e-06
UniRef100_UPI0000E203DD PREDICTED: mucin 7, salivary isoform 2 n... 56 2e-06
UniRef100_A1XYV9 Latency-associated nuclear antigen n=1 Tax=Retr... 56 2e-06
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 56 2e-06
UniRef100_C0VX39 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 56 2e-06
UniRef100_Q9LV48 Protein kinase-like protein n=1 Tax=Arabidopsis... 56 2e-06
UniRef100_Q6QJ28 Putative uncharacterized protein (Fragment) n=1... 56 2e-06
UniRef100_C5Z794 Putative uncharacterized protein Sb10g026240 n=... 56 2e-06
UniRef100_C0PPC0 Putative uncharacterized protein n=1 Tax=Zea ma... 56 2e-06
UniRef100_B9G127 Putative uncharacterized protein n=1 Tax=Oryza ... 56 2e-06
UniRef100_B6KAY2 Putative uncharacterized protein n=2 Tax=Toxopl... 56 2e-06
UniRef100_Q86ZG7 Probable Cytokinesis protein sepA n=1 Tax=Neuro... 56 2e-06
UniRef100_Q6CEK4 YALI0B14971p n=1 Tax=Yarrowia lipolytica RepID=... 56 2e-06
UniRef100_C9SGQ7 Putative uncharacterized protein n=1 Tax=Vertic... 56 2e-06
UniRef100_B8M4W4 Actin associated protein Wsp1, putative n=1 Tax... 56 2e-06
UniRef100_B8M4W3 Actin associated protein Wsp1, putative n=1 Tax... 56 2e-06
UniRef100_Q89893 Uncharacterized protein DR2 n=1 Tax=Human herpe... 56 2e-06
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI00017F04CF PREDICTED: similar to Uncharacterized pr... 55 2e-06
UniRef100_UPI00017974A0 PREDICTED: similar to WAS/WASL interacti... 55 2e-06
UniRef100_UPI00015550FC PREDICTED: similar to scavenger receptor... 55 2e-06
UniRef100_UPI0000E1F7FF PREDICTED: similar to WASPIP protein iso... 55 2e-06
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 55 2e-06
UniRef100_UPI0000D9EBC8 PREDICTED: similar to gametogenetin n=1 ... 55 2e-06
UniRef100_UPI00005EA817 PREDICTED: similar to SH3 domain-binding... 55 2e-06
UniRef100_UPI00017B2A81 UPI00017B2A81 related cluster n=1 Tax=Te... 55 2e-06
UniRef100_A3Q0W3 Putative uncharacterized protein n=1 Tax=Mycoba... 55 2e-06
UniRef100_C6RBK9 Iron-sulfur cluster-binding protein n=1 Tax=Cor... 55 2e-06
UniRef100_C2CRU3 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 55 2e-06
UniRef100_C0WGM5 Fe-S oxidoreductase n=1 Tax=Corynebacterium acc... 55 2e-06
UniRef100_Q9FPQ5 Gamete-specific hydroxyproline-rich glycoprotei... 55 2e-06
UniRef100_Q94JZ6 At3g24600 n=1 Tax=Arabidopsis thaliana RepID=Q9... 55 2e-06
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 55 2e-06
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 55 2e-06
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_A9SZ15 Non-specific lipid-transfer protein n=2 Tax=Phy... 55 2e-06
UniRef100_A8JIL0 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 55 2e-06
UniRef100_Q86HN1 Putative uncharacterized protein gxcX n=1 Tax=D... 55 2e-06
UniRef100_Q4FX64 Proteophosphoglycan ppg3, putative n=1 Tax=Leis... 55 2e-06
UniRef100_Q55RM6 Putative uncharacterized protein n=1 Tax=Filoba... 55 2e-06
UniRef100_C4JT61 Predicted protein n=1 Tax=Uncinocarpus reesii 1... 55 2e-06
UniRef100_C0NIF7 Putative uncharacterized protein n=1 Tax=Ajello... 55 2e-06
UniRef100_A6ZPV2 Conserved protein n=1 Tax=Saccharomyces cerevis... 55 2e-06
UniRef100_A6RH13 Predicted protein n=1 Tax=Ajellomyces capsulatu... 55 2e-06
UniRef100_A1DF14 Cell wall protein, putative n=1 Tax=Neosartorya... 55 2e-06
UniRef100_A1D2Q3 Conserved proline-rich protein n=1 Tax=Neosarto... 55 2e-06
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 55 2e-06
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 55 2e-06
UniRef100_UPI000175F960 PREDICTED: similar to WAS/WASL interacti... 55 3e-06
UniRef100_UPI00016A4DAA hypothetical protein BoklE_07101 n=1 Tax... 55 3e-06
UniRef100_UPI0000E12B23 Os07g0496300 n=1 Tax=Oryza sativa Japoni... 55 3e-06
UniRef100_UPI00005A1EA0 PREDICTED: similar to Bile-salt-activate... 55 3e-06
UniRef100_UPI00005A1E9F PREDICTED: similar to Bile-salt-activate... 55 3e-06
UniRef100_UPI00005A1E9E PREDICTED: similar to Bile-salt-activate... 55 3e-06
UniRef100_UPI00005A1E9D PREDICTED: similar to Bile-salt-activate... 55 3e-06
UniRef100_UPI0001A2D7F0 WAS/WASL interacting protein family memb... 55 3e-06
UniRef100_UPI0000EB2391 Bile salt-activated lipase precursor (EC... 55 3e-06
UniRef100_UPI0000EB1A8B UPI0000EB1A8B related cluster n=1 Tax=Ca... 55 3e-06
UniRef100_Q65553 UL36 n=3 Tax=Bovine herpesvirus 1 RepID=Q65553_... 55 3e-06
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 55 3e-06
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 55 3e-06
UniRef100_Q6YY67 Putative uncharacterized protein OSJNBa0006O15.... 55 3e-06
UniRef100_Q5VR94 Putative uncharacterized protein P0038C05.36 n=... 55 3e-06
UniRef100_Q0WWI8 Early nodulin-like 2 predicted GPI-anchored pro... 55 3e-06
UniRef100_C5XD41 Putative uncharacterized protein Sb02g037970 n=... 55 3e-06
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 55 3e-06
UniRef100_A8JKC9 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 55 3e-06
UniRef100_Q08DX4 WAS/WASL interacting protein family, member 1 n... 55 3e-06
UniRef100_Q9Y075 Proteophosphoglycan (Fragment) n=1 Tax=Leishman... 55 3e-06
UniRef100_Q7PIC2 AGAP006340-PA n=1 Tax=Anopheles gambiae RepID=Q... 55 3e-06
UniRef100_Q5CKR5 Putative uncharacterized protein n=1 Tax=Crypto... 55 3e-06
UniRef100_Q4D9L5 Putative uncharacterized protein n=1 Tax=Trypan... 55 3e-06
UniRef100_A8JUX2 Rab3-GEF, isoform B n=1 Tax=Drosophila melanoga... 55 3e-06
UniRef100_A7UU74 AGAP006340-PC n=1 Tax=Anopheles gambiae RepID=A... 55 3e-06
UniRef100_A7UU73 AGAP006340-PB n=1 Tax=Anopheles gambiae RepID=A... 55 3e-06
UniRef100_Q96VI4 Protease 1 n=1 Tax=Pneumocystis carinii RepID=Q... 55 3e-06
UniRef100_Q6CBU0 YALI0C15532p n=1 Tax=Yarrowia lipolytica RepID=... 55 3e-06
UniRef100_Q07229 Verprolin n=1 Tax=Saccharomyces cerevisiae RepI... 55 3e-06
UniRef100_C9SV27 Pyruvate dehydrogenase protein X component n=1 ... 55 3e-06
UniRef100_C5MHK9 Predicted protein n=1 Tax=Candida tropicalis MY... 55 3e-06
UniRef100_C5FF74 Splicing factor 3B subunit 4 n=1 Tax=Microsporu... 55 3e-06
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 55 3e-06
UniRef100_C0RYB1 Putative uncharacterized protein n=1 Tax=Paraco... 55 3e-06
UniRef100_B2VSJ8 Putative uncharacterized protein n=1 Tax=Pyreno... 55 3e-06
UniRef100_A2R0A5 Remark: C-terminal truncated ORF due to end of ... 55 3e-06
UniRef100_O43516-4 Isoform 4 of WAS/WASL-interacting protein fam... 55 3e-06
UniRef100_P37370 Verprolin n=1 Tax=Saccharomyces cerevisiae RepI... 55 3e-06
UniRef100_Q9VXY2 MAP kinase-activating death domain protein n=1 ... 55 3e-06
UniRef100_UPI0000F3115A fms-related tyrosine kinase 3 ligand n=2... 55 4e-06
UniRef100_UPI0000D9B0F8 PREDICTED: similar to mucin 7, salivary ... 55 4e-06
UniRef100_UPI00006A21F8 UPI00006A21F8 related cluster n=1 Tax=Xe... 55 4e-06
UniRef100_UPI00006A0BFF Nuclear receptor coactivator 1 (EC 2.3.1... 55 4e-06
UniRef100_Q5F3Q7 Putative uncharacterized protein n=1 Tax=Gallus... 55 4e-06
UniRef100_Q4RQ25 Chromosome 17 SCAF15006, whole genome shotgun s... 55 4e-06
UniRef100_B4UEI7 2-alkenal reductase n=1 Tax=Anaeromyxobacter sp... 55 4e-06
UniRef100_A8IEX2 Putative uncharacterized protein n=1 Tax=Azorhi... 55 4e-06
UniRef100_A0LSH8 Glycoside hydrolase, family 6 n=1 Tax=Acidother... 55 4e-06
UniRef100_Q9SKM6 En/Spm-like transposon protein n=1 Tax=Arabidop... 55 4e-06
UniRef100_Q7XNA5 OSJNBa0011E07.13 protein n=1 Tax=Oryza sativa J... 55 4e-06
UniRef100_Q6Z1Z8 Os08g0100500 protein n=1 Tax=Oryza sativa Japon... 55 4e-06
UniRef100_Q6Z1Z7 cDNA clone:001-206-C02, full insert sequence n=... 55 4e-06
UniRef100_C5Y027 Putative uncharacterized protein Sb04g010730 n=... 55 4e-06
UniRef100_C1FJU1 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 4e-06
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_B9FTC9 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_B8B9X3 Putative uncharacterized protein n=1 Tax=Oryza ... 55 4e-06
UniRef100_B6U4I2 Putative uncharacterized protein n=1 Tax=Zea ma... 55 4e-06
UniRef100_B6U1N0 Putative uncharacterized protein n=1 Tax=Zea ma... 55 4e-06
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 55 4e-06
UniRef100_B9PVZ4 Putative uncharacterized protein n=1 Tax=Toxopl... 55 4e-06
UniRef100_A4HCC8 Putative uncharacterized protein n=1 Tax=Leishm... 55 4e-06
UniRef100_Q7SFQ1 Predicted protein n=1 Tax=Neurospora crassa Rep... 55 4e-06
UniRef100_Q6AHT0 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 55 4e-06
UniRef100_Q4WUE6 Cell wall protein, putative n=1 Tax=Aspergillus... 55 4e-06
UniRef100_Q0D1Z1 Putative uncharacterized protein n=1 Tax=Asperg... 55 4e-06
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 55 4e-06
UniRef100_C5M739 Predicted protein n=1 Tax=Candida tropicalis MY... 55 4e-06
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 55 4e-06
UniRef100_C1GKW7 Predicted protein n=1 Tax=Paracoccidioides bras... 55 4e-06
UniRef100_B8LVW5 Proline-rich, actin-associated protein Vrp1, pu... 55 4e-06
UniRef100_B0Y3X0 Cell wall protein, putative n=1 Tax=Aspergillus... 55 4e-06
UniRef100_A1CPW0 Conserved proline-rich protein n=1 Tax=Aspergil... 55 4e-06
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 55 4e-06
UniRef100_UPI0001985C7D PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_UPI00017F0583 PREDICTED: similar to Ras and Rab intera... 54 5e-06
UniRef100_UPI0000D9D097 PREDICTED: similar to Wiskott-Aldrich sy... 54 5e-06
UniRef100_UPI0000D9D096 PREDICTED: similar to Wiskott-Aldrich sy... 54 5e-06
UniRef100_UPI0000D9D095 PREDICTED: similar to Wiskott-Aldrich sy... 54 5e-06
UniRef100_UPI0000D9D094 PREDICTED: similar to Wiskott-Aldrich sy... 54 5e-06
UniRef100_UPI0000EB3858 UPI0000EB3858 related cluster n=1 Tax=Ca... 54 5e-06
UniRef100_Q3UKQ3 Putative uncharacterized protein n=1 Tax=Mus mu... 54 5e-06
UniRef100_B2RYB3 Srrm1 protein n=1 Tax=Rattus norvegicus RepID=B... 54 5e-06
UniRef100_A8L8X7 Membrane-bound lytic murein transglycosylase B-... 54 5e-06
UniRef100_Q9SX31 F24J5.8 protein n=1 Tax=Arabidopsis thaliana Re... 54 5e-06
UniRef100_Q4KXE0 Cold acclimation induced protein 2-1 n=1 Tax=Tr... 54 5e-06
UniRef100_C5YBI1 Putative uncharacterized protein Sb06g021550 n=... 54 5e-06
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 54 5e-06
UniRef100_B8B912 Putative uncharacterized protein n=1 Tax=Oryza ... 54 5e-06
UniRef100_Q28659 Fertilin alpha subunit n=1 Tax=Oryctolagus cuni... 54 5e-06
UniRef100_Q5CHL1 Hydroxyproline-rich glycoprotein dz-hrgp n=1 Ta... 54 5e-06
UniRef100_Q28Z79 GA21415 n=1 Tax=Drosophila pseudoobscura pseudo... 54 5e-06
UniRef100_B4IQK0 GM19288 n=1 Tax=Drosophila sechellia RepID=B4IQ... 54 5e-06
UniRef100_B4IMI5 GM16182 n=1 Tax=Drosophila sechellia RepID=B4IM... 54 5e-06
UniRef100_B4GI56 GL17658 n=1 Tax=Drosophila persimilis RepID=B4G... 54 5e-06
UniRef100_A7SQQ5 Predicted protein n=1 Tax=Nematostella vectensi... 54 5e-06
UniRef100_A3FPW7 Formin-related protein, putative n=2 Tax=Crypto... 54 5e-06
UniRef100_Q96VJ1 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 54 5e-06
UniRef100_Q0UMH1 Putative uncharacterized protein n=1 Tax=Phaeos... 54 5e-06
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 54 5e-06
UniRef100_B3LQ87 Myosin tail region-interacting protein MTI1 n=1... 54 5e-06
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 54 5e-06
UniRef100_UPI0001982E0B PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI000175F600 PREDICTED: hypothetical protein n=1 Tax=... 54 7e-06
UniRef100_UPI0001555C17 PREDICTED: hypothetical protein, partial... 54 7e-06
UniRef100_Q8BZD7 Putative uncharacterized protein (Fragment) n=2... 54 7e-06
UniRef100_Q3U6Q1 Putative uncharacterized protein n=1 Tax=Mus mu... 54 7e-06
UniRef100_Q3T9Z7 Putative uncharacterized protein n=1 Tax=Mus mu... 54 7e-06
UniRef100_Q096E0 FrgA n=1 Tax=Stigmatella aurantiaca DW4/3-1 Rep... 54 7e-06
UniRef100_B4VBA7 Putative uncharacterized protein n=1 Tax=Strept... 54 7e-06
UniRef100_Q9CAL8 Putative uncharacterized protein F24J13.3 n=1 T... 54 7e-06
UniRef100_Q5JNC9 Os01g0738300 protein n=1 Tax=Oryza sativa Japon... 54 7e-06
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 54 7e-06
UniRef100_B9EZL1 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_B8A933 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_A8MRQ5 Uncharacterized protein At4g22485.1 n=1 Tax=Ara... 54 7e-06
UniRef100_A8IZG7 Plus agglutinin protein (Fragment) n=1 Tax=Chla... 54 7e-06
UniRef100_A7QW77 Chromosome chr3 scaffold_199, whole genome shot... 54 7e-06
UniRef100_A2XD25 Putative uncharacterized protein n=1 Tax=Oryza ... 54 7e-06
UniRef100_B9QJU2 Putative uncharacterized protein n=1 Tax=Toxopl... 54 7e-06
UniRef100_B9PLD5 Putative uncharacterized protein n=1 Tax=Toxopl... 54 7e-06
UniRef100_B4IN27 GM17403 n=1 Tax=Drosophila sechellia RepID=B4IN... 54 7e-06
UniRef100_Q5KG31 Putative uncharacterized protein n=1 Tax=Filoba... 54 7e-06
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 54 7e-06
UniRef100_Q1DV84 Putative uncharacterized protein n=1 Tax=Coccid... 54 7e-06
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 54 7e-06
UniRef100_C5JCP6 Proline-rich n=1 Tax=Ajellomyces dermatitidis S... 54 7e-06
UniRef100_C5GW70 Proline-rich n=1 Tax=Ajellomyces dermatitidis E... 54 7e-06
UniRef100_A7TP17 Putative uncharacterized protein n=1 Tax=Vander... 54 7e-06
UniRef100_Q923D5 WW domain-binding protein 11 n=1 Tax=Mus muscul... 54 7e-06
UniRef100_Q8BH43 Wiskott-Aldrich syndrome protein family member ... 54 7e-06
UniRef100_O15047 Histone-lysine N-methyltransferase SETD1A n=1 T... 54 7e-06
UniRef100_UPI00017390C7 AGP19 (ARABINOGALACTAN-PROTEIN 19) n=1 T... 54 9e-06
UniRef100_UPI00015B42DB PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI00015562F3 PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI000151B498 hypothetical protein PGUG_04978 n=1 Tax=... 54 9e-06
UniRef100_UPI0000F2E24E PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI0000DD9486 Os08g0280200 n=1 Tax=Oryza sativa Japoni... 54 9e-06
UniRef100_UPI0000D9EA10 PREDICTED: hypothetical protein n=1 Tax=... 54 9e-06
UniRef100_UPI000036F39A PREDICTED: hypothetical protein isoform ... 54 9e-06
UniRef100_UPI000023DB3E hypothetical protein FG02559.1 n=1 Tax=G... 54 9e-06
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 54 9e-06
UniRef100_Q1QHE7 OmpA/MotB n=1 Tax=Nitrobacter hamburgensis X14 ... 54 9e-06
UniRef100_Q8GX23 Putative serine/threonine protein kinase n=1 Ta... 54 9e-06
UniRef100_Q67W44 Os06g0486000 protein n=1 Tax=Oryza sativa Japon... 54 9e-06
UniRef100_Q4KXD9 Early salt stress and cold acclimation-induced ... 54 9e-06
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 54 9e-06
UniRef100_O65672 Putative serine/threonine protein kinase n=1 Ta... 54 9e-06
UniRef100_B8B2F0 Putative uncharacterized protein n=1 Tax=Oryza ... 54 9e-06
UniRef100_A9U3I4 Predicted protein n=1 Tax=Physcomitrella patens... 54 9e-06
UniRef100_C3ZT83 Putative uncharacterized protein n=1 Tax=Branch... 54 9e-06
UniRef100_B6KLQ3 Putative uncharacterized protein n=1 Tax=Toxopl... 54 9e-06
UniRef100_B6ACL5 Putative uncharacterized protein n=1 Tax=Crypto... 54 9e-06
UniRef100_Q2U304 Predicted protein n=1 Tax=Aspergillus oryzae Re... 54 9e-06
UniRef100_C5E045 ZYRO0G09680p n=1 Tax=Zygosaccharomyces rouxii C... 54 9e-06
UniRef100_B6QR64 Proline-rich, actin-associated protein Vrp1, pu... 54 9e-06
UniRef100_B2AAE4 Predicted CDS Pa_1_3720 n=1 Tax=Podospora anser... 54 9e-06
UniRef100_A5DNX7 Putative uncharacterized protein n=1 Tax=Pichia... 54 9e-06
UniRef100_Q9Y2W2 WW domain-binding protein 11 n=1 Tax=Homo sapie... 54 9e-06
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 54 9e-06
UniRef100_Q6ZCX3 Formin-like protein 6 n=1 Tax=Oryza sativa Japo... 54 9e-06
UniRef100_Q9C6S1 Formin-like protein 14 n=1 Tax=Arabidopsis thal... 54 9e-06
UniRef100_Q9S740 Lysine-rich arabinogalactan protein 19 n=1 Tax=... 54 9e-06