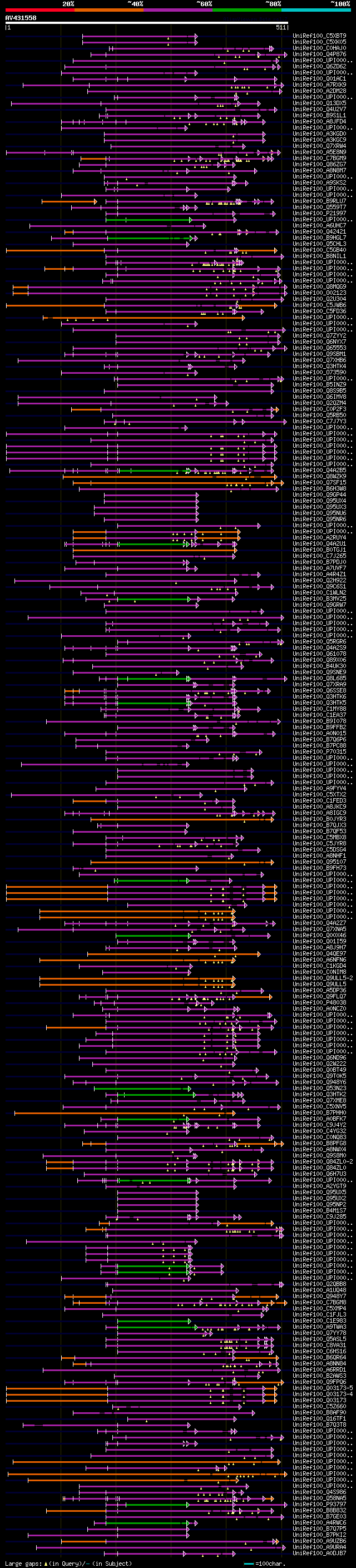
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV431558 PM002c08_r
(511 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_C5XBT9 Putative uncharacterized protein Sb02g005130 n=... 73 1e-11
UniRef100_C5XK05 Putative uncharacterized protein Sb03g034303 (F... 72 2e-11
UniRef100_C0HAJ0 Wiskott-Aldrich syndrome protein homolog n=1 Ta... 70 6e-11
UniRef100_Q4P876 Putative uncharacterized protein n=1 Tax=Ustila... 70 6e-11
UniRef100_UPI0001B55E13 putative chaplin n=1 Tax=Streptomyces sp... 70 1e-10
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 68 3e-10
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 68 4e-10
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 68 4e-10
UniRef100_A7RXK9 Predicted protein n=1 Tax=Nematostella vectensi... 67 5e-10
UniRef100_A2DM28 Diaphanous, putative n=1 Tax=Trichomonas vagina... 67 5e-10
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 67 7e-10
UniRef100_Q13DX5 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Bi... 67 7e-10
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 67 7e-10
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 67 7e-10
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 67 7e-10
UniRef100_UPI000179DCA7 Zinc finger homeodomain 4 n=1 Tax=Bos ta... 67 9e-10
UniRef100_A3KGD0 Asparagine-linked glycosylation 13 homolog (S. ... 67 9e-10
UniRef100_A3KGC9 Asparagine-linked glycosylation 13 homolog (S. ... 67 9e-10
UniRef100_Q7XRW4 OSJNBb0062H02.5 protein n=2 Tax=Oryza sativa Re... 67 9e-10
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 66 1e-09
UniRef100_C7BGM9 Formin 2B n=1 Tax=Physcomitrella patens RepID=C... 66 1e-09
UniRef100_Q86ZG7 Probable Cytokinesis protein sepA n=1 Tax=Neuro... 66 1e-09
UniRef100_A8N8M7 Putative uncharacterized protein n=1 Tax=Coprin... 66 1e-09
UniRef100_UPI0000DC1C18 UPI0000DC1C18 related cluster n=1 Tax=Ra... 66 2e-09
UniRef100_A9SKS2 Predicted protein n=1 Tax=Physcomitrella patens... 66 2e-09
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 66 2e-09
UniRef100_UPI00006A0DA0 zinc finger homeodomain 4 n=1 Tax=Xenopu... 66 2e-09
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 66 2e-09
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 66 2e-09
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 66 2e-09
UniRef100_UPI0000DB78D4 PREDICTED: hypothetical protein n=1 Tax=... 65 2e-09
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 65 2e-09
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 65 2e-09
UniRef100_B9HGL7 Predicted protein n=1 Tax=Populus trichocarpa R... 65 2e-09
UniRef100_Q5CHL3 Hydroxyproline-rich glycoprotein dz-hrgp n=1 Ta... 65 2e-09
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 65 2e-09
UniRef100_B8NIL1 Proline-rich, actin-associated protein Vrp1, pu... 65 2e-09
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 65 3e-09
UniRef100_UPI0000DB6FAC PREDICTED: similar to Protein cappuccino... 65 3e-09
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 65 3e-09
UniRef100_Q8MQG9 Prion-like-(Q/n-rich)-domain-bearing protein pr... 65 3e-09
UniRef100_O02123 Prion-like-(Q/n-rich)-domain-bearing protein pr... 65 3e-09
UniRef100_Q2U304 Predicted protein n=1 Tax=Aspergillus oryzae Re... 65 3e-09
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 65 3e-09
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 65 3e-09
UniRef100_UPI0000F2CB25 PREDICTED: similar to HOMER-3A (flip) pr... 65 3e-09
UniRef100_UPI0000E7FF28 PREDICTED: similar to zinc-finger homeod... 65 3e-09
UniRef100_UPI0000DA45C5 PREDICTED: similar to diacylglycerol kin... 65 3e-09
UniRef100_Q7ZYY2 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 65 3e-09
UniRef100_Q6NYX7 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 65 3e-09
UniRef100_Q65553 UL36 n=3 Tax=Bovine herpesvirus 1 RepID=Q65553_... 65 3e-09
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 65 3e-09
UniRef100_Q7XHB6 Transposon protein, putative, CACTA, En/Spm sub... 65 3e-09
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 65 3e-09
UniRef100_O73590 Zinc finger homeobox protein 4 n=1 Tax=Gallus g... 65 3e-09
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 64 5e-09
UniRef100_B5INZ9 Penicillin-binding protein 1A n=1 Tax=Cyanobium... 64 5e-09
UniRef100_Q8S9B5 Matrix metalloproteinase n=1 Tax=Volvox carteri... 64 5e-09
UniRef100_Q6IMV8 Transposase n=1 Tax=Oryza sativa Indica Group R... 64 5e-09
UniRef100_Q2QZM4 Transposon protein, putative, CACTA, En/Spm sub... 64 5e-09
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 64 5e-09
UniRef100_Q5RB50 Putative uncharacterized protein DKFZp459N037 n... 64 5e-09
UniRef100_C7J7Y3 Os10g0324900 protein (Fragment) n=1 Tax=Oryza s... 64 6e-09
UniRef100_UPI000194C052 PREDICTED: dishevelled associated activa... 64 6e-09
UniRef100_UPI0000F53460 enabled homolog isoform 2 n=2 Tax=Mus mu... 64 6e-09
UniRef100_UPI00005A2B62 PREDICTED: similar to Wiskott-Aldrich sy... 64 6e-09
UniRef100_UPI000047625B enabled homolog isoform 3 n=1 Tax=Mus mu... 64 6e-09
UniRef100_UPI00015DF6E3 enabled homolog (Drosophila) n=1 Tax=Mus... 64 6e-09
UniRef100_UPI000056479E enabled homolog isoform 1 n=1 Tax=Mus mu... 64 6e-09
UniRef100_UPI00005A2B61 PREDICTED: similar to Wiskott-Aldrich sy... 64 6e-09
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 64 6e-09
UniRef100_Q8WZK9 Putative uncharacterized protein B14D6.190 n=2 ... 64 6e-09
UniRef100_Q7SF15 WASP-like pretein las17p n=1 Tax=Neurospora cra... 64 6e-09
UniRef100_B6H3W8 Pc13g11020 protein n=1 Tax=Penicillium chrysoge... 64 6e-09
UniRef100_Q9GP44 NONA protein (Fragment) n=1 Tax=Drosophila viri... 64 8e-09
UniRef100_Q95UX4 No on or off transient A (Fragment) n=1 Tax=Dro... 64 8e-09
UniRef100_Q95UX3 No on or off transient A (Fragment) n=1 Tax=Dro... 64 8e-09
UniRef100_Q95NU6 No on or off transient A (Fragment) n=1 Tax=Dro... 64 8e-09
UniRef100_Q95NR6 No on or off transient A (Fragment) n=1 Tax=Dro... 64 8e-09
UniRef100_UPI000023DD8E hypothetical protein FG06945.1 n=1 Tax=G... 64 8e-09
UniRef100_UPI0001A2BBDD hypothetical protein LOC100009637 n=1 Ta... 64 8e-09
UniRef100_A2RUY4 Zgc:158395 protein n=1 Tax=Danio rerio RepID=A2... 64 8e-09
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 64 8e-09
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 64 8e-09
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 64 8e-09
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 64 8e-09
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 64 8e-09
UniRef100_A4R4Z1 Putative uncharacterized protein n=1 Tax=Magnap... 64 8e-09
UniRef100_Q2H922 Actin cytoskeleton-regulatory complex protein P... 64 8e-09
UniRef100_Q9C6S1 Formin-like protein 14 n=1 Tax=Arabidopsis thal... 64 8e-09
UniRef100_C1WLN2 Putative uncharacterized protein n=1 Tax=Kribbe... 63 1e-08
UniRef100_B3MV25 GF22854 n=1 Tax=Drosophila ananassae RepID=B3MV... 63 1e-08
UniRef100_Q9GRW7 Protein no-on-transient A n=1 Tax=Drosophila vi... 63 1e-08
UniRef100_UPI000194C983 PREDICTED: inverted formin, FH2 and WH2 ... 63 1e-08
UniRef100_UPI0000E80E27 PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI0000D9B853 PREDICTED: similar to Formin-1 isoform I... 63 1e-08
UniRef100_UPI00016E98C2 UPI00016E98C2 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_UPI0000ECD10B Zinc finger homeobox protein 4 (Zinc fin... 63 1e-08
UniRef100_Q5RGR6 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 63 1e-08
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 63 1e-08
UniRef100_Q61078 Wiscott-Aldrich Syndrome protein homolog n=1 Ta... 63 1e-08
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 63 1e-08
UniRef100_B4UK30 TonB family protein n=1 Tax=Anaeromyxobacter sp... 63 1e-08
UniRef100_Q9SNE9 Putative uncharacterized protein F11C1_20 n=1 T... 63 1e-08
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 63 1e-08
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 63 1e-08
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 63 1e-08
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 63 1e-08
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 63 1e-08
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 63 1e-08
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 63 1e-08
UniRef100_B9I078 Predicted protein n=1 Tax=Populus trichocarpa R... 63 1e-08
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 63 1e-08
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 63 1e-08
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 63 1e-08
UniRef100_P70315 Wiskott-Aldrich syndrome protein homolog n=2 Ta... 63 1e-08
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI00005EB969 PREDICTED: similar to SET binding protei... 63 1e-08
UniRef100_UPI00016E10B5 UPI00016E10B5 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_UPI00016E10B4 UPI00016E10B4 related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_UPI0000E7F7FF PREDICTED: similar to N-WASP n=1 Tax=Gal... 63 1e-08
UniRef100_A9FYV4 Putative uncharacterized protein n=1 Tax=Sorang... 63 1e-08
UniRef100_C5XTX2 Putative uncharacterized protein Sb04g002820 n=... 63 1e-08
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 63 1e-08
UniRef100_A8JKC9 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 63 1e-08
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 63 1e-08
UniRef100_B0JYR3 Wiskott-Aldrich syndrome-like n=1 Tax=Bos tauru... 63 1e-08
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 63 1e-08
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 63 1e-08
UniRef100_C5MBX8 Putative uncharacterized protein n=1 Tax=Candid... 63 1e-08
UniRef100_C5JYR8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 63 1e-08
UniRef100_C5DSG4 ZYRO0B16654p n=1 Tax=Zygosaccharomyces rouxii C... 63 1e-08
UniRef100_A8NHF1 Predicted protein n=1 Tax=Coprinopsis cinerea o... 63 1e-08
UniRef100_Q95107 Neural Wiskott-Aldrich syndrome protein n=1 Tax... 63 1e-08
UniRef100_B9FH73 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 62 2e-08
UniRef100_UPI0000DA3EC9 PREDICTED: similar to tumor endothelial ... 62 2e-08
UniRef100_UPI0001B7A6BD enabled homolog n=1 Tax=Rattus norvegicu... 62 2e-08
UniRef100_UPI0001B7A6BC enabled homolog n=1 Tax=Rattus norvegicu... 62 2e-08
UniRef100_UPI0001B7A6A7 enabled homolog n=1 Tax=Rattus norvegicu... 62 2e-08
UniRef100_UPI00015DEC3D chromodomain helicase DNA binding protei... 62 2e-08
UniRef100_UPI00015DF9DB Proline-rich protein 12. n=1 Tax=Homo sa... 62 2e-08
UniRef100_UPI0001596889 proline rich 12 n=1 Tax=Homo sapiens Rep... 62 2e-08
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 62 2e-08
UniRef100_Q7XNA5 OSJNBa0011E07.13 protein n=1 Tax=Oryza sativa J... 62 2e-08
UniRef100_Q00X46 Chromosome 13 contig 1, DNA sequence n=1 Tax=Os... 62 2e-08
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 62 2e-08
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 62 2e-08
UniRef100_Q4QE97 Formin A n=1 Tax=Leishmania major RepID=Q4QE97_... 62 2e-08
UniRef100_A6NFN6 Putative uncharacterized protein PRR12 n=1 Tax=... 62 2e-08
UniRef100_C1KGD4 SepA/Bni1 n=1 Tax=Epichloe festucae RepID=C1KGD... 62 2e-08
UniRef100_C0NIM8 Putative uncharacterized protein n=1 Tax=Ajello... 62 2e-08
UniRef100_Q9ULL5-2 Isoform 2 of Proline-rich protein 12 n=1 Tax=... 62 2e-08
UniRef100_Q9ULL5 Proline-rich protein 12 n=1 Tax=Homo sapiens Re... 62 2e-08
UniRef100_A5DP36 Actin cytoskeleton-regulatory complex protein P... 62 2e-08
UniRef100_Q9FLQ7 Formin-like protein 20 n=1 Tax=Arabidopsis thal... 62 2e-08
UniRef100_P48038 Acrosin heavy chain n=1 Tax=Oryctolagus cunicul... 62 2e-08
UniRef100_A0NCZ0 AGAP001055-PA (Fragment) n=1 Tax=Anopheles gamb... 62 2e-08
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 62 2e-08
UniRef100_UPI0000E800CF PREDICTED: similar to formin 2 n=1 Tax=G... 62 2e-08
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 62 2e-08
UniRef100_UPI00016E900E UPI00016E900E related cluster n=1 Tax=Ta... 62 2e-08
UniRef100_UPI00016E900D UPI00016E900D related cluster n=1 Tax=Ta... 62 2e-08
UniRef100_UPI00016E900C UPI00016E900C related cluster n=1 Tax=Ta... 62 2e-08
UniRef100_UPI0000ECC8C2 Formin-2. n=1 Tax=Gallus gallus RepID=UP... 62 2e-08
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 62 2e-08
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 62 2e-08
UniRef100_Q0BT49 Hypothetical secreted protein n=1 Tax=Granuliba... 62 2e-08
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 62 2e-08
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 62 2e-08
UniRef100_Q53N23 Transposon protein, putative, CACTA, En/Spm sub... 62 2e-08
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 62 2e-08
UniRef100_Q7XME8 OSJNBa0061G20.11 protein n=2 Tax=Oryza sativa R... 62 2e-08
UniRef100_C5XNV5 Putative uncharacterized protein Sb03g026090 n=... 62 2e-08
UniRef100_B7PHH0 Cyclin t, putative n=1 Tax=Ixodes scapularis Re... 62 2e-08
UniRef100_A0BFK7 Chromosome undetermined scaffold_104, whole gen... 62 2e-08
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 62 2e-08
UniRef100_C4YG32 Putative uncharacterized protein n=1 Tax=Candid... 62 2e-08
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 62 2e-08
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 62 2e-08
UniRef100_A8NWX4 Putative uncharacterized protein n=1 Tax=Coprin... 62 2e-08
UniRef100_Q9S8M0 Chitin-binding lectin 1 n=1 Tax=Solanum tuberos... 62 2e-08
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 62 2e-08
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 62 2e-08
UniRef100_Q6H7U3 Formin-like protein 10 n=1 Tax=Oryza sativa Jap... 62 2e-08
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 62 3e-08
UniRef100_A2YGT9 Putative uncharacterized protein n=1 Tax=Oryza ... 62 3e-08
UniRef100_Q95UX5 No on or off transient A (Fragment) n=1 Tax=Dro... 62 3e-08
UniRef100_Q95UX2 No on or off transient A (Fragment) n=1 Tax=Dro... 62 3e-08
UniRef100_Q95NP2 No on or off transient A (Fragment) n=1 Tax=Dro... 62 3e-08
UniRef100_B4M1S7 NonA n=1 Tax=Drosophila virilis RepID=B4M1S7_DROVI 62 3e-08
UniRef100_C9J285 Putative uncharacterized protein ENSP0000039240... 62 3e-08
UniRef100_UPI000151AF18 hypothetical protein PGUG_03629 n=1 Tax=... 62 3e-08
UniRef100_UPI0000E48C31 PREDICTED: hypothetical protein n=1 Tax=... 62 3e-08
UniRef100_UPI0000DAC50E ubiquitin E3 ligase ICP0 n=1 Tax=Papiine... 62 3e-08
UniRef100_UPI000025003E PREDICTED: similar to Homeobox protein e... 62 3e-08
UniRef100_UPI00016E72E0 UPI00016E72E0 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E72DE UPI00016E72DE related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E72DD UPI00016E72DD related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E10B6 UPI00016E10B6 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI00016E10A1 UPI00016E10A1 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_UPI000184A096 Zinc finger homeobox protein 4 (Zinc fin... 62 3e-08
UniRef100_Q2QBB8 RL2 n=1 Tax=Papiine herpesvirus 2 RepID=Q2QBB8_... 62 3e-08
UniRef100_A1UQ48 Peptidase S8 and S53, subtilisin, kexin, sedoli... 62 3e-08
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 62 3e-08
UniRef100_C7BGM8 Formin 2A n=1 Tax=Physcomitrella patens RepID=C... 62 3e-08
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 62 3e-08
UniRef100_C1FJL3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 62 3e-08
UniRef100_C1E983 Predicted protein n=1 Tax=Micromonas sp. RCC299... 62 3e-08
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 62 3e-08
UniRef100_Q7YY78 Protease, possible n=2 Tax=Cryptosporidium parv... 62 3e-08
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 62 3e-08
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 62 3e-08
UniRef100_C6HS16 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 62 3e-08
UniRef100_B6QR64 Proline-rich, actin-associated protein Vrp1, pu... 62 3e-08
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 62 3e-08
UniRef100_A6RRD1 Putative uncharacterized protein n=1 Tax=Botryo... 62 3e-08
UniRef100_B2AWS3 Actin cytoskeleton-regulatory complex protein P... 62 3e-08
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 62 3e-08
UniRef100_Q03173-5 Isoform 4 of Protein enabled homolog n=1 Tax=... 62 3e-08
UniRef100_Q03173-4 Isoform 3 of Protein enabled homolog n=1 Tax=... 62 3e-08
UniRef100_Q03173 Protein enabled homolog n=1 Tax=Mus musculus Re... 62 3e-08
UniRef100_C5Z660 Putative uncharacterized protein Sb10g024410 n=... 61 4e-08
UniRef100_B8AF90 Putative uncharacterized protein n=1 Tax=Oryza ... 61 4e-08
UniRef100_Q16TF1 Fibrillarin (Fragment) n=1 Tax=Aedes aegypti Re... 61 4e-08
UniRef100_B7Q3T8 Putative uncharacterized protein (Fragment) n=1... 61 4e-08
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 61 4e-08
UniRef100_UPI000194E282 PREDICTED: similar to Wiskott-Aldrich sy... 61 4e-08
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 61 4e-08
UniRef100_UPI0001796B35 PREDICTED: formin-like 1 n=1 Tax=Equus c... 61 4e-08
UniRef100_UPI0001795F41 PREDICTED: similar to WASL protein n=1 T... 61 4e-08
UniRef100_UPI0000F2C402 PREDICTED: similar to voltage-gated calc... 61 4e-08
UniRef100_UPI0000ECB813 Formin-like protein 2 (Formin homology 2... 61 4e-08
UniRef100_UPI0000E7FF26 PREDICTED: similar to MSC protein n=1 Ta... 61 4e-08
UniRef100_UPI0000E48A44 PREDICTED: hypothetical protein n=1 Tax=... 61 4e-08
UniRef100_UPI00016E8FE9 UPI00016E8FE9 related cluster n=1 Tax=Ta... 61 4e-08
UniRef100_Q4S986 Chromosome 3 SCAF14700, whole genome shotgun se... 61 4e-08
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 61 4e-08
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 61 4e-08
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 61 4e-08
UniRef100_B7GE03 Predicted protein n=1 Tax=Phaeodactylum tricorn... 61 4e-08
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 61 4e-08
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 61 4e-08
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 61 4e-08
UniRef100_A9UZB6 Predicted protein n=1 Tax=Monosiga brevicollis ... 61 4e-08
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 61 4e-08
UniRef100_A0DJB7 Chromosome undetermined scaffold_53, whole geno... 61 4e-08
UniRef100_A0BLV2 Chromosome undetermined scaffold_115, whole gen... 61 4e-08
UniRef100_Q2U9G0 Rho GTPase effector BNI1 and related formins n=... 61 4e-08
UniRef100_Q2H4B7 Predicted protein n=1 Tax=Chaetomium globosum R... 61 4e-08
UniRef100_C9S8M2 Cytokinesis protein sepA n=1 Tax=Verticillium a... 61 4e-08
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 61 4e-08
UniRef100_C5FH48 Basic proline-rich protein n=1 Tax=Microsporum ... 61 4e-08
UniRef100_B8ND33 Cytokinesis protein SepA n=1 Tax=Aspergillus fl... 61 4e-08
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 61 4e-08
UniRef100_UPI0000E4617D PREDICTED: similar to Activating signal ... 61 5e-08
UniRef100_UPI0000DA3D7B PREDICTED: hypothetical protein n=1 Tax=... 61 5e-08
UniRef100_C1MP90 Predicted protein n=1 Tax=Micromonas pusilla CC... 61 5e-08
UniRef100_UPI000179366E PREDICTED: similar to CG32138 CG32138-PB... 61 5e-08
UniRef100_UPI000151B9E8 hypothetical protein PGUG_05037 n=1 Tax=... 61 5e-08
UniRef100_UPI0000DABDCC ubiquitin E3 ligase ICP0 n=1 Tax=Macacin... 61 5e-08
UniRef100_UPI0000DA267E Wiskott-Aldrich syndrome-like n=1 Tax=Ra... 61 5e-08
UniRef100_UPI00005A2F6D PREDICTED: similar to CG33556-PA n=2 Tax... 61 5e-08
UniRef100_A2VDK6 Wiskott-Aldrich syndrome protein family member ... 61 5e-08
UniRef100_UPI0000EC9F55 Anti-Mullerian hormone n=1 Tax=Gallus ga... 61 5e-08
UniRef100_Q7T400 Immediate early protein ICP0 n=1 Tax=Macacine h... 61 5e-08
UniRef100_Q2J3C8 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Ha... 61 5e-08
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 61 5e-08
UniRef100_Q9LUI1 At3g22800 n=1 Tax=Arabidopsis thaliana RepID=Q9... 61 5e-08
UniRef100_Q6Z1Z8 Os08g0100500 protein n=1 Tax=Oryza sativa Japon... 61 5e-08
UniRef100_Q6Z1Z7 cDNA clone:001-206-C02, full insert sequence n=... 61 5e-08
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 61 5e-08
UniRef100_Q00TD0 Chromosome 17 contig 1, DNA sequence. (Fragment... 61 5e-08
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 61 5e-08
UniRef100_C0PLL4 Putative uncharacterized protein n=1 Tax=Zea ma... 61 5e-08
UniRef100_B9MXQ3 Predicted protein (Fragment) n=1 Tax=Populus tr... 61 5e-08
UniRef100_B9FY87 Putative uncharacterized protein n=1 Tax=Oryza ... 61 5e-08
UniRef100_B8B9X3 Putative uncharacterized protein n=1 Tax=Oryza ... 61 5e-08
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 61 5e-08
UniRef100_Q5CLH8 Protease n=1 Tax=Cryptosporidium hominis RepID=... 61 5e-08
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 61 5e-08
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 61 5e-08
UniRef100_B5DZ38 GA24855 n=1 Tax=Drosophila pseudoobscura pseudo... 61 5e-08
UniRef100_A9V3V4 Predicted protein n=1 Tax=Monosiga brevicollis ... 61 5e-08
UniRef100_Q7RWH7 Cytokinesis protein sepA n=1 Tax=Neurospora cra... 61 5e-08
UniRef100_Q1E016 Predicted protein n=1 Tax=Coccidioides immitis ... 61 5e-08
UniRef100_C4YB97 Putative uncharacterized protein n=1 Tax=Clavis... 61 5e-08
UniRef100_B9WMX2 Actin assembly factor, putative n=1 Tax=Candida... 61 5e-08
UniRef100_B0CYD2 WASP-interacting protein VRP1/WIP n=1 Tax=Lacca... 61 5e-08
UniRef100_O08816 Neural Wiskott-Aldrich syndrome protein n=1 Tax... 61 5e-08
UniRef100_UPI0000F2DD8B PREDICTED: similar to zinc finger protei... 60 6e-08
UniRef100_UPI0000DA1BD4 PREDICTED: hypothetical protein n=1 Tax=... 60 6e-08
UniRef100_Q1XG61 Putative glycine-rich RNA binding protein n=1 T... 60 6e-08
UniRef100_O62006 Intermediate filament protein C2 n=1 Tax=Branch... 60 6e-08
UniRef100_B4L7Q3 GI11213 n=1 Tax=Drosophila mojavensis RepID=B4L... 60 6e-08
UniRef100_A6RGJ8 Predicted protein n=1 Tax=Ajellomyces capsulatu... 60 6e-08
UniRef100_Q9HE26 rRNA 2'-O-methyltransferase fibrillarin n=1 Tax... 60 6e-08
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 60 7e-08
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 60 7e-08
UniRef100_UPI0000E4896A PREDICTED: similar to CG33556-PA n=1 Tax... 60 7e-08
UniRef100_UPI000023F096 hypothetical protein FG08656.1 n=1 Tax=G... 60 7e-08
UniRef100_Q0RAQ2 Putative uncharacterized protein n=1 Tax=Franki... 60 7e-08
UniRef100_B1VR69 Putative uncharacterized protein n=1 Tax=Strept... 60 7e-08
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 60 7e-08
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 7e-08
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 60 7e-08
UniRef100_B8ATX9 Putative uncharacterized protein n=1 Tax=Oryza ... 60 7e-08
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 60 7e-08
UniRef100_A8J3Q3 Glyoxal or galactose oxidase n=1 Tax=Chlamydomo... 60 7e-08
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 60 7e-08
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 60 7e-08
UniRef100_B4KHD1 GI15288 n=1 Tax=Drosophila mojavensis RepID=B4K... 60 7e-08
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 60 7e-08
UniRef100_A9V6R1 Predicted protein n=1 Tax=Monosiga brevicollis ... 60 7e-08
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 60 7e-08
UniRef100_Q17R91 DIAPH2 protein n=1 Tax=Homo sapiens RepID=Q17R9... 60 7e-08
UniRef100_C9J6U3 Putative uncharacterized protein DIAPH2 n=1 Tax... 60 7e-08
UniRef100_B7ZLJ0 DIAPH2 protein n=1 Tax=Homo sapiens RepID=B7ZLJ... 60 7e-08
UniRef100_B3KMJ1 cDNA FLJ11167 fis, clone PLACE1007257, highly s... 60 7e-08
UniRef100_A8MSQ4 Diaphanous homolog 2 (Drosophila), isoform CRA_... 60 7e-08
UniRef100_A8K5F7 cDNA FLJ75405, highly similar to Homo sapiens d... 60 7e-08
UniRef100_A6NML8 Diaphanous homolog 2 (Drosophila), isoform CRA_... 60 7e-08
UniRef100_C5DK76 KLTH0F02376p n=1 Tax=Lachancea thermotolerans C... 60 7e-08
UniRef100_C4JK96 Putative uncharacterized protein n=1 Tax=Uncino... 60 7e-08
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 60 7e-08
UniRef100_O60879-2 Isoform 2 of Protein diaphanous homolog 2 n=1... 60 7e-08
UniRef100_O60879-3 Isoform 3 of Protein diaphanous homolog 2 n=1... 60 7e-08
UniRef100_O60879 Protein diaphanous homolog 2 n=1 Tax=Homo sapie... 60 7e-08
UniRef100_UPI0000DB75B1 PREDICTED: similar to One cut domain fam... 60 8e-08
UniRef100_UPI0000DA32FB PREDICTED: hypothetical protein n=1 Tax=... 60 8e-08
UniRef100_UPI0000DA1CBA PREDICTED: hypothetical protein n=1 Tax=... 60 8e-08
UniRef100_UPI0000DBF1EE UPI0000DBF1EE related cluster n=1 Tax=Ra... 60 8e-08
UniRef100_C1MWG0 Predicted protein n=1 Tax=Micromonas pusilla CC... 60 8e-08
UniRef100_Q95UW8 No on or off transient A (Fragment) n=1 Tax=Dro... 60 8e-08
UniRef100_Q95UW1 No on or off transient A (Fragment) n=1 Tax=Dro... 60 8e-08
UniRef100_UPI0000F2C630 PREDICTED: similar to zinc finger homeod... 60 9e-08
UniRef100_UPI0000D9ED26 PREDICTED: hypothetical protein n=1 Tax=... 60 9e-08
UniRef100_UPI0000D9EC9F PREDICTED: similar to Myb protein P42POP... 60 9e-08
UniRef100_UPI00004D6F7F Formin-like protein 2 (Formin homology 2... 60 9e-08
UniRef100_UPI0001B798A6 UPI0001B798A6 related cluster n=1 Tax=Ho... 60 9e-08
UniRef100_UPI000179CCA4 UPI000179CCA4 related cluster n=1 Tax=Bo... 60 9e-08
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 60 9e-08
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 60 9e-08
UniRef100_B8AQT2 Putative uncharacterized protein n=1 Tax=Oryza ... 60 9e-08
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 60 9e-08
UniRef100_A8HU70 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 60 9e-08
UniRef100_A7QQ26 Chromosome chr2 scaffold_140, whole genome shot... 60 9e-08
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 60 9e-08
UniRef100_A9VAA9 Predicted protein n=1 Tax=Monosiga brevicollis ... 60 9e-08
UniRef100_A0DA74 Chromosome undetermined scaffold_43, whole geno... 60 9e-08
UniRef100_Q55S44 Putative uncharacterized protein n=1 Tax=Filoba... 60 9e-08
UniRef100_Q4WCV2 Actin associated protein Wsp1, putative n=1 Tax... 60 9e-08
UniRef100_C5P6Q6 WH1 domain containing protein n=1 Tax=Coccidioi... 60 9e-08
UniRef100_B9W9S7 Formin (Bud-site selection/polarity protein), p... 60 9e-08
UniRef100_A4RL74 Putative uncharacterized protein n=1 Tax=Magnap... 60 9e-08
UniRef100_A4QPP9 Putative uncharacterized protein n=1 Tax=Magnap... 60 9e-08
UniRef100_Q86VE0 Myb-related transcription factor, partner of pr... 60 9e-08
UniRef100_Q96PY5-3 Isoform 2 of Formin-like protein 2 n=1 Tax=Ho... 60 9e-08
UniRef100_Q96PY5 Formin-like protein 2 n=1 Tax=Homo sapiens RepI... 60 9e-08
UniRef100_UPI00005A3429 PREDICTED: similar to hemojuvelin isofor... 60 1e-07
UniRef100_A5GPV8 RNA-binding protein, RRM domain n=1 Tax=Synecho... 60 1e-07
UniRef100_A5GHM5 RNA-binding protein, RRM domain n=1 Tax=Synecho... 60 1e-07
UniRef100_O65514 Putative glycine-rich cell wall protein n=1 Tax... 60 1e-07
UniRef100_C1N3K2 Predicted protein n=1 Tax=Micromonas pusilla CC... 60 1e-07
UniRef100_UPI000198316D PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI0001982977 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI00017F09BE PREDICTED: similar to KIAA1902 protein, ... 60 1e-07
UniRef100_UPI00017974C7 PREDICTED: similar to KIAA1902 protein n... 60 1e-07
UniRef100_UPI0001796908 PREDICTED: zinc finger homeobox 4 n=1 Ta... 60 1e-07
UniRef100_UPI0000F2E472 PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI0000E20F55 PREDICTED: dishevelled associated activa... 60 1e-07
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 60 1e-07
UniRef100_UPI0000D9ACBC PREDICTED: dishevelled associated activa... 60 1e-07
UniRef100_UPI0000231CFE dishevelled associated activator of morp... 60 1e-07
UniRef100_UPI000069FBE3 Formin-like protein 2 (Formin homology 2... 60 1e-07
UniRef100_UPI000069FBE2 Formin-like protein 2 (Formin homology 2... 60 1e-07
UniRef100_Q6NRE3 MGC83886 protein n=1 Tax=Xenopus laevis RepID=Q... 60 1e-07
UniRef100_Q4RQM1 Chromosome 2 SCAF15004, whole genome shotgun se... 60 1e-07
UniRef100_Q829E0 Putative membrane protein n=1 Tax=Streptomyces ... 60 1e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 60 1e-07
UniRef100_B8J593 TonB family protein n=1 Tax=Anaeromyxobacter de... 60 1e-07
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 60 1e-07
UniRef100_Q2R0D0 Transposon protein, putative, CACTA, En/Spm sub... 60 1e-07
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 60 1e-07
UniRef100_C5X5H7 Putative uncharacterized protein Sb02g042640 n=... 60 1e-07
UniRef100_C1EC31 Predicted protein n=1 Tax=Micromonas sp. RCC299... 60 1e-07
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 60 1e-07
UniRef100_B9RFA2 Putative uncharacterized protein n=1 Tax=Ricinu... 60 1e-07
UniRef100_B9H5S2 Predicted protein n=1 Tax=Populus trichocarpa R... 60 1e-07
UniRef100_B8A1H5 Putative uncharacterized protein n=1 Tax=Zea ma... 60 1e-07
UniRef100_B4FW52 Putative uncharacterized protein n=1 Tax=Zea ma... 60 1e-07
UniRef100_A8HTD7 Glyoxal or galactose oxidase n=1 Tax=Chlamydomo... 60 1e-07
UniRef100_Q9GSG9 Wiscott-Aldrich syndrome protein n=1 Tax=Dictyo... 60 1e-07
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 60 1e-07
UniRef100_B7P3M5 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_A0D1C0 Chromosome undetermined scaffold_34, whole geno... 60 1e-07
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 60 1e-07
UniRef100_C9K0J5 Putative uncharacterized protein RAPH1 n=1 Tax=... 60 1e-07
UniRef100_Q5KAA5 Cytokinesis protein sepa (Fh1/2 protein), putat... 60 1e-07
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 60 1e-07
UniRef100_C5GLX8 Cytokinesis protein sepA n=1 Tax=Ajellomyces de... 60 1e-07
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 60 1e-07
UniRef100_C5DNK9 KLTH0G17886p n=1 Tax=Lachancea thermotolerans C... 60 1e-07
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 60 1e-07
UniRef100_C0NKI4 Basic proline-rich protein n=1 Tax=Ajellomyces ... 60 1e-07
UniRef100_B8PC49 Predicted protein n=1 Tax=Postia placenta Mad-6... 60 1e-07
UniRef100_B0D333 Predicted protein n=1 Tax=Laccaria bicolor S238... 60 1e-07
UniRef100_A6QWU5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 60 1e-07
UniRef100_A5DRR5 Putative uncharacterized protein n=1 Tax=Lodder... 60 1e-07
UniRef100_Q70E73 Ras-associated and pleckstrin homology domains-... 60 1e-07
UniRef100_Q86T65 Disheveled-associated activator of morphogenesi... 60 1e-07
UniRef100_UPI0001925556 PREDICTED: similar to Putative RNA-bindi... 59 1e-07
UniRef100_UPI0000E4981C PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_Q7TWC0 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 59 1e-07
UniRef100_Q6MWW7 PE-PGRS FAMILY PROTEIN (Fragment) n=1 Tax=Mycob... 59 1e-07
UniRef100_Q3KEU2 Putative glycine rich protein n=1 Tax=Pseudomon... 59 1e-07
UniRef100_Q3B0Y9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 59 1e-07
UniRef100_Q5Z8T9 Os06g0706700 protein n=1 Tax=Oryza sativa Japon... 59 1e-07
UniRef100_Q9GP77 NONA protein (Fragment) n=1 Tax=Drosophila litt... 59 1e-07
UniRef100_Q95UW6 No on or off transient A (Fragment) n=1 Tax=Dro... 59 1e-07
UniRef100_Q95UW5 No on or off transient A (Fragment) n=1 Tax=Dro... 59 1e-07
UniRef100_Q5TU53 AGAP002965-PA (Fragment) n=1 Tax=Anopheles gamb... 59 1e-07
UniRef100_B7QN34 Putative uncharacterized protein (Fragment) n=1... 59 1e-07
UniRef100_B7PCR0 Cement protein, putative (Fragment) n=1 Tax=Ixo... 59 1e-07
UniRef100_Q0RDS4 Translation initiation factor IF-2 n=1 Tax=Fran... 59 1e-07
UniRef100_UPI0001A7B186 actin binding n=1 Tax=Arabidopsis thalia... 59 1e-07
UniRef100_UPI00019261C6 PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_UPI0000F2E754 PREDICTED: similar to hCG38312, n=1 Tax=... 59 1e-07
UniRef100_UPI0000E81081 PREDICTED: similar to G protein-coupled ... 59 1e-07
UniRef100_UPI0000E80087 PREDICTED: similar to RP1-278E11.1 n=1 T... 59 1e-07
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 59 1e-07
UniRef100_UPI0000E1F761 PREDICTED: formin-like 2 isoform 2 n=1 T... 59 1e-07
UniRef100_UPI0000E1F760 PREDICTED: formin-like 2 isoform 8 n=1 T... 59 1e-07
UniRef100_UPI00005A5941 PREDICTED: similar to Wiskott-Aldrich sy... 59 1e-07
UniRef100_UPI0000DC1294 UPI0000DC1294 related cluster n=1 Tax=Ra... 59 1e-07
UniRef100_UPI00016E72DF UPI00016E72DF related cluster n=1 Tax=Ta... 59 1e-07
UniRef100_UPI0001951250 Formin-like protein 2 (Formin homology 2... 59 1e-07
UniRef100_UPI0000EB01B2 WAS/WASL interacting protein family memb... 59 1e-07
UniRef100_UPI0000ECC879 Disheveled-associated activator of morph... 59 1e-07
UniRef100_UPI0000ECC878 Disheveled-associated activator of morph... 59 1e-07
UniRef100_Q91810 Proline rich protein n=1 Tax=Xenopus laevis Rep... 59 1e-07
UniRef100_Q640C2 LOC397922 protein n=1 Tax=Xenopus laevis RepID=... 59 1e-07
UniRef100_Q4TB69 Chromosome 11 SCAF7190, whole genome shotgun se... 59 1e-07
UniRef100_Q4RE14 Chromosome undetermined SCAF15155, whole genome... 59 1e-07
UniRef100_Q7XSN2 OSJNBb0028M18.5 protein n=1 Tax=Oryza sativa Ja... 59 1e-07
UniRef100_Q5VR02 Hydroxyproline-rich glycoprotein-like n=1 Tax=O... 59 1e-07
UniRef100_Q2QVU2 Transposon protein, putative, CACTA, En/Spm sub... 59 1e-07
UniRef100_Q01MU7 OSIGBa0102O13.9 protein n=1 Tax=Oryza sativa Re... 59 1e-07
UniRef100_C1MSQ5 Predicted protein n=1 Tax=Micromonas pusilla CC... 59 1e-07
UniRef100_A8JBZ3 Metalloproteinase of VMP family n=1 Tax=Chlamyd... 59 1e-07
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 59 1e-07
UniRef100_A7XQ02 Latex protein n=1 Tax=Morus alba RepID=A7XQ02_M... 59 1e-07
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 59 1e-07
UniRef100_B5DI49 GA25909 n=1 Tax=Drosophila pseudoobscura pseudo... 59 1e-07
UniRef100_B4NYZ4 GE18300 n=1 Tax=Drosophila yakuba RepID=B4NYZ4_... 59 1e-07
UniRef100_B4G8I0 GL19302 n=1 Tax=Drosophila persimilis RepID=B4G... 59 1e-07
UniRef100_A5JUU8 Formin B n=2 Tax=Trypanosoma brucei RepID=A5JUU... 59 1e-07
UniRef100_A0D550 Chromosome undetermined scaffold_38, whole geno... 59 1e-07
UniRef100_Q2KFY0 Putative uncharacterized protein n=1 Tax=Magnap... 59 1e-07
UniRef100_Q2HG22 Putative uncharacterized protein n=1 Tax=Chaeto... 59 1e-07
UniRef100_B8LVW5 Proline-rich, actin-associated protein Vrp1, pu... 59 1e-07
UniRef100_B2B5R8 Predicted CDS Pa_2_5560 (Fragment) n=1 Tax=Podo... 59 1e-07
UniRef100_A5DK28 Putative uncharacterized protein n=1 Tax=Pichia... 59 1e-07
UniRef100_A4RBS7 Putative uncharacterized protein n=1 Tax=Magnap... 59 1e-07
UniRef100_Q9LVN1 Formin-like protein 13 n=1 Tax=Arabidopsis thal... 59 1e-07
UniRef100_UPI0000F2E7F3 PREDICTED: similar to twist transcriptio... 59 2e-07
UniRef100_UPI0000DA2377 PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_UPI0000DA1EB9 PREDICTED: hypothetical protein n=1 Tax=... 59 2e-07
UniRef100_UPI0000DBF4CE UPI0000DBF4CE related cluster n=1 Tax=Ra... 59 2e-07
UniRef100_C1YHF7 Membrane carboxypeptidase (Penicillin-binding p... 59 2e-07
UniRef100_A6GBL5 RNA-binding protein n=1 Tax=Plesiocystis pacifi... 59 2e-07
UniRef100_B9NDK1 Predicted protein n=1 Tax=Populus trichocarpa R... 59 2e-07
UniRef100_B7QNG2 Glycine-rich protein, putative (Fragment) n=1 T... 59 2e-07
UniRef100_UPI0001AF1AE4 hypothetical protein SghaA1_22823 n=1 Ta... 59 2e-07
UniRef100_UPI000194C22E PREDICTED: similar to formin 2 n=1 Tax=T... 59 2e-07
UniRef100_UPI00017C2CD7 PREDICTED: similar to formin-like 2 n=1 ... 59 2e-07
UniRef100_UPI0001795BE7 PREDICTED: similar to FH2 domain contain... 59 2e-07
UniRef100_UPI000175F21B PREDICTED: similar to Ras-associated and... 59 2e-07
UniRef100_UPI0000E48DD8 PREDICTED: similar to MGC81512 protein n... 59 2e-07
UniRef100_UPI0000E1276E Os06g0280500 n=1 Tax=Oryza sativa Japoni... 59 2e-07
UniRef100_UPI0000E12293 Os03g0809400 n=1 Tax=Oryza sativa Japoni... 59 2e-07
UniRef100_UPI00015A6C38 UPI00015A6C38 related cluster n=1 Tax=Da... 59 2e-07
UniRef100_UPI00017B188D UPI00017B188D related cluster n=1 Tax=Te... 59 2e-07
UniRef100_UPI00001809D7 UPI00001809D7 related cluster n=1 Tax=Ra... 59 2e-07
UniRef100_UPI0001AE68B4 UPI0001AE68B4 related cluster n=1 Tax=Ho... 59 2e-07
UniRef100_UPI00016E8FEB UPI00016E8FEB related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_UPI00016E8720 UPI00016E8720 related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_UPI00016E871F UPI00016E871F related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 59 2e-07
UniRef100_UPI000179F39E UPI000179F39E related cluster n=1 Tax=Bo... 59 2e-07
UniRef100_Q6DD43 LOC446221 protein (Fragment) n=1 Tax=Xenopus la... 59 2e-07
UniRef100_Q5ID03 Enabled protein n=1 Tax=Xenopus laevis RepID=Q5... 59 2e-07
UniRef100_Q4V859 LOC446221 protein n=1 Tax=Xenopus laevis RepID=... 59 2e-07
UniRef100_B0R0R1 Novel protein similar to human Ras association ... 59 2e-07
UniRef100_Q7TPN5 Wiskott-Aldrich syndrome-like (Human) n=2 Tax=M... 59 2e-07
UniRef100_Q5FWU0 WAS protein family, member 2 n=1 Tax=Rattus nor... 59 2e-07
UniRef100_C5C089 Metallophosphoesterase n=1 Tax=Beutenbergia cav... 59 2e-07