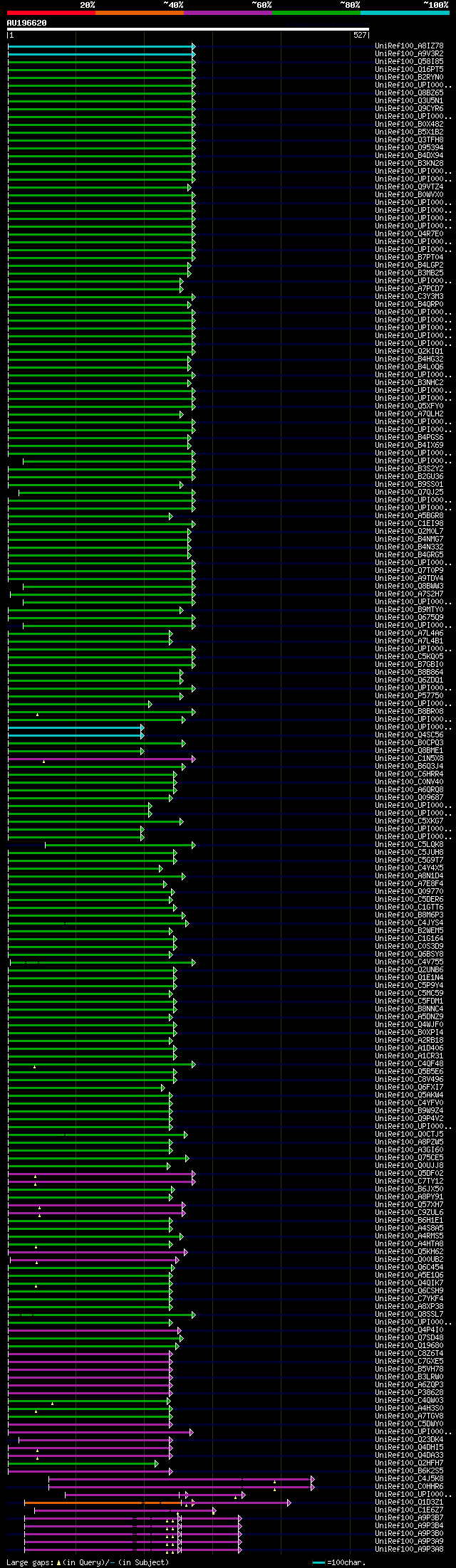
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AU196620 PFL092c01_r
(527 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_A8IZ78 Predicted protein n=1 Tax=Chlamydomonas reinhar... 178 2e-43
UniRef100_A9V3R2 Predicted protein n=1 Tax=Monosiga brevicollis ... 127 4e-28
UniRef100_Q58I85 Phosphoacetylglucosamine mutase n=1 Tax=Aedes a... 125 2e-27
UniRef100_Q16PT5 Phosphoglucomutase n=1 Tax=Aedes aegypti RepID=... 125 2e-27
UniRef100_B2RYN0 Pgm3 protein n=1 Tax=Rattus norvegicus RepID=B2... 119 1e-25
UniRef100_UPI000155EDCF PREDICTED: similar to Phosphoacetylgluco... 119 1e-25
UniRef100_Q8BZ65 Putative uncharacterized protein n=1 Tax=Mus mu... 117 4e-25
UniRef100_Q3U5N1 Putative uncharacterized protein (Fragment) n=1... 117 4e-25
UniRef100_Q9CYR6 Phosphoacetylglucosamine mutase n=1 Tax=Mus mus... 117 4e-25
UniRef100_UPI0000D5707F PREDICTED: similar to phosphoacetylgluco... 117 5e-25
UniRef100_B0X482 Phosphoglucomutase n=1 Tax=Culex quinquefasciat... 117 5e-25
UniRef100_B5X1B2 Phosphoacetylglucosamine mutase n=1 Tax=Salmo s... 117 6e-25
UniRef100_Q3TFH8 Putative uncharacterized protein n=1 Tax=Mus mu... 117 6e-25
UniRef100_O95394 Phosphoacetylglucosamine mutase n=2 Tax=Homo sa... 116 8e-25
UniRef100_B4DX94 cDNA FLJ55543, highly similar to Phosphoacetylg... 116 8e-25
UniRef100_B3KN28 cDNA FLJ13370 fis, clone PLACE1000653, highly s... 116 8e-25
UniRef100_UPI0001864A9F hypothetical protein BRAFLDRAFT_124253 n... 115 2e-24
UniRef100_UPI00016E8E33 UPI00016E8E33 related cluster n=1 Tax=Ta... 115 2e-24
UniRef100_Q9VTZ4 CG10627 n=1 Tax=Drosophila melanogaster RepID=Q... 115 2e-24
UniRef100_B0WVX0 Phosphoglucomutase n=1 Tax=Culex quinquefasciat... 115 2e-24
UniRef100_UPI0000E2105E PREDICTED: phosphoglucomutase 3 isoform ... 114 3e-24
UniRef100_UPI0000D9ADA3 PREDICTED: phosphoglucomutase 3 isoform ... 114 3e-24
UniRef100_UPI0000D9ADA2 PREDICTED: phosphoglucomutase 3 isoform ... 114 3e-24
UniRef100_UPI000036D886 PREDICTED: phosphoglucomutase 3 isoform ... 114 3e-24
UniRef100_Q4R7E0 Testis cDNA, clone: QtsA-15546, similar to huma... 114 3e-24
UniRef100_UPI00015B59BD PREDICTED: similar to phosphoglucomutase... 114 4e-24
UniRef100_UPI00017F0A9C PREDICTED: similar to Phosphoglucomutase... 114 5e-24
UniRef100_B7PT04 Phosphoacetylglucosamine mutase, putative (Frag... 114 5e-24
UniRef100_B4LGP2 GJ13809 n=1 Tax=Drosophila virilis RepID=B4LGP2... 114 5e-24
UniRef100_B3MB25 GF24006 n=1 Tax=Drosophila ananassae RepID=B3MB... 114 5e-24
UniRef100_UPI0001982C75 PREDICTED: hypothetical protein isoform ... 113 7e-24
UniRef100_A7PCD7 Chromosome chr2 scaffold_11, whole genome shotg... 113 7e-24
UniRef100_C3Y3M3 Putative uncharacterized protein n=1 Tax=Branch... 113 7e-24
UniRef100_B4QRP0 GD12708 n=1 Tax=Drosophila simulans RepID=B4QRP... 112 1e-23
UniRef100_UPI00005A27AA PREDICTED: similar to Phosphoacetylgluco... 112 2e-23
UniRef100_UPI00005A27A9 PREDICTED: similar to Phosphoacetylgluco... 112 2e-23
UniRef100_UPI00005A27A8 PREDICTED: similar to Phosphoacetylgluco... 112 2e-23
UniRef100_UPI00004A55BC PREDICTED: similar to Phosphoacetylgluco... 112 2e-23
UniRef100_UPI00005BD423 hypothetical protein LOC505648 n=1 Tax=B... 112 2e-23
UniRef100_Q2KIQ1 Phosphoglucomutase 3 n=1 Tax=Bos taurus RepID=Q... 112 2e-23
UniRef100_B4HG32 GM24644 n=1 Tax=Drosophila sechellia RepID=B4HG... 111 3e-23
UniRef100_B4L0Q6 GI13061 n=1 Tax=Drosophila mojavensis RepID=B4L... 111 4e-23
UniRef100_UPI000155CF87 PREDICTED: similar to N-acetylglucosamin... 110 5e-23
UniRef100_B3NHC2 GG13815 n=1 Tax=Drosophila erecta RepID=B3NHC2_... 110 5e-23
UniRef100_UPI0000F2C1F5 PREDICTED: similar to N-acetylglucosamin... 110 8e-23
UniRef100_UPI0001A2D8D6 phosphoglucomutase 3 n=1 Tax=Danio rerio... 110 8e-23
UniRef100_Q5XFY0 Phosphoglucomutase 3 n=1 Tax=Danio rerio RepID=... 110 8e-23
UniRef100_A7QLH2 Chromosome undetermined scaffold_119, whole gen... 110 8e-23
UniRef100_UPI00003AD07C PREDICTED: similar to N-acetylglucosamin... 109 1e-22
UniRef100_UPI00006108A1 Phosphoacetylglucosamine mutase (EC 5.4.... 109 1e-22
UniRef100_B4PGS6 GE20109 n=1 Tax=Drosophila yakuba RepID=B4PGS6_... 108 2e-22
UniRef100_B4IX69 GH16842 n=1 Tax=Drosophila grimshawi RepID=B4IX... 108 2e-22
UniRef100_UPI000180B33F PREDICTED: similar to phosphoglucomutase... 108 3e-22
UniRef100_UPI000021F5D1 UPI000021F5D1 related cluster n=1 Tax=Ra... 107 5e-22
UniRef100_B3S2Y2 Putative uncharacterized protein n=1 Tax=Tricho... 107 5e-22
UniRef100_B2GU36 LOC100158504 protein n=1 Tax=Xenopus (Silurana)... 107 7e-22
UniRef100_B9SS01 Phosphoglucomutase, putative n=1 Tax=Ricinus co... 107 7e-22
UniRef100_Q7QJ25 AGAP007215-PA n=1 Tax=Anopheles gambiae RepID=Q... 107 7e-22
UniRef100_UPI0001792CCF PREDICTED: similar to phosphoglucomutase... 106 9e-22
UniRef100_UPI00004D1C1D Phosphoacetylglucosamine mutase (EC 5.4.... 106 9e-22
UniRef100_A5BGR8 Putative uncharacterized protein n=1 Tax=Vitis ... 106 9e-22
UniRef100_C1EI98 Predicted protein n=1 Tax=Micromonas sp. RCC299... 106 1e-21
UniRef100_Q2M0L7 GA10449 n=1 Tax=Drosophila pseudoobscura pseudo... 106 1e-21
UniRef100_B4NMG7 GK23090 n=1 Tax=Drosophila willistoni RepID=B4N... 106 1e-21
UniRef100_B4N332 GK12616 n=1 Tax=Drosophila willistoni RepID=B4N... 106 1e-21
UniRef100_B4GRG5 GL24942 n=1 Tax=Drosophila persimilis RepID=B4G... 106 1e-21
UniRef100_UPI000194C0E4 PREDICTED: hypothetical protein n=1 Tax=... 105 1e-21
UniRef100_Q7T0P9 Pgm3-prov protein n=1 Tax=Xenopus laevis RepID=... 105 1e-21
UniRef100_A9TDV4 Predicted protein n=1 Tax=Physcomitrella patens... 105 1e-21
UniRef100_Q8BWW3 Putative uncharacterized protein n=1 Tax=Mus mu... 105 2e-21
UniRef100_A7S2H7 Predicted protein n=1 Tax=Nematostella vectensi... 105 3e-21
UniRef100_UPI00016E8E34 UPI00016E8E34 related cluster n=1 Tax=Ta... 103 7e-21
UniRef100_B9MTY0 Predicted protein n=1 Tax=Populus trichocarpa R... 102 2e-20
UniRef100_Q675Q9 Phosphoacetylglucosamine mutase n=1 Tax=Oikople... 101 4e-20
UniRef100_UPI00004BBD0B PREDICTED: similar to Phosphoacetylgluco... 100 8e-20
UniRef100_A7L4A6 Phosphoglucosamine mutase n=1 Tax=Carica papaya... 99 2e-19
UniRef100_A7L4B1 Phosphoglucosamine mutase n=1 Tax=Carica papaya... 99 2e-19
UniRef100_UPI0000DB7504 PREDICTED: similar to CG10627-PA n=1 Tax... 98 4e-19
UniRef100_C5KQ05 Phosphoacetylglucosamine mutase, putative n=1 T... 98 4e-19
UniRef100_B7GBI0 Predicted protein n=1 Tax=Phaeodactylum tricorn... 97 7e-19
UniRef100_B8B864 Putative uncharacterized protein n=1 Tax=Oryza ... 96 1e-18
UniRef100_Q6ZDQ1 Phosphoacetylglucosamine mutase n=2 Tax=Oryza s... 95 3e-18
UniRef100_UPI000186EE58 Phosphoacetylglucosamine mutase, putativ... 94 8e-18
UniRef100_P57750 Phosphoacetylglucosamine mutase n=1 Tax=Arabido... 94 8e-18
UniRef100_UPI000154E5DC phosphoglucomutase 3 n=1 Tax=Rattus norv... 92 2e-17
UniRef100_B8BR08 Putative uncharacterized protein (Fragment) n=1... 92 3e-17
UniRef100_UPI000187C400 hypothetical protein MPER_00735 n=1 Tax=... 91 4e-17
UniRef100_UPI00017B170D UPI00017B170D related cluster n=1 Tax=Te... 90 8e-17
UniRef100_Q4SC56 Chromosome 14 SCAF14660, whole genome shotgun s... 90 8e-17
UniRef100_B0CPQ3 Phosphoacetylglucosamine mutase n=1 Tax=Laccari... 90 1e-16
UniRef100_Q8BME1 Putative uncharacterized protein n=1 Tax=Mus mu... 89 1e-16
UniRef100_C1N5X8 Predicted protein n=1 Tax=Micromonas pusilla CC... 89 2e-16
UniRef100_B6Q3J4 N-acetylglucosamine-phosphate mutase n=1 Tax=Pe... 88 3e-16
UniRef100_C6HRR4 N-acetylglucosamine-phosphate mutase n=1 Tax=Aj... 87 7e-16
UniRef100_C0NV40 N-acetylglucosamine-phosphate mutase n=1 Tax=Aj... 87 7e-16
UniRef100_A6QRQ8 Putative uncharacterized protein n=1 Tax=Ajello... 87 7e-16
UniRef100_Q09687 Probable phosphoacetylglucosamine mutase 1 n=1 ... 87 7e-16
UniRef100_UPI0000E21060 PREDICTED: phosphoglucomutase 3 isoform ... 87 9e-16
UniRef100_UPI0000D9ADA5 PREDICTED: phosphoglucomutase 3 isoform ... 87 9e-16
UniRef100_C5XKG7 Putative uncharacterized protein Sb03g001710 n=... 87 9e-16
UniRef100_UPI0000E2105F PREDICTED: phosphoglucomutase 3 isoform ... 86 2e-15
UniRef100_UPI0000D9ADA4 PREDICTED: phosphoglucomutase 3 isoform ... 86 2e-15
UniRef100_C5LQK8 Phosphoglucomutase, putative (Fragment) n=1 Tax... 86 2e-15
UniRef100_C5JUH8 N-acetylglucosamine-phosphate mutase n=1 Tax=Aj... 86 2e-15
UniRef100_C5G9T7 N-acetylglucosamine-phosphate mutase n=1 Tax=Aj... 86 2e-15
UniRef100_C4Y4X5 Putative uncharacterized protein n=1 Tax=Clavis... 86 2e-15
UniRef100_A8N1D4 Putative uncharacterized protein n=1 Tax=Coprin... 86 2e-15
UniRef100_A7E8F4 Putative uncharacterized protein n=1 Tax=Sclero... 85 3e-15
UniRef100_Q09770 Probable phosphoacetylglucosamine mutase 2 n=1 ... 85 3e-15
UniRef100_C5DER6 KLTH0C11506p n=1 Tax=Lachancea thermotolerans C... 85 4e-15
UniRef100_C1GTT6 Phosphoacetylglucosamine mutase n=1 Tax=Paracoc... 85 4e-15
UniRef100_B8M6P3 N-acetylglucosamine-phosphate mutase n=1 Tax=Ta... 84 6e-15
UniRef100_C4JYS4 Phosphoacetylglucosamine mutase n=1 Tax=Uncinoc... 84 8e-15
UniRef100_B2WEM5 N-acetylglucosamine-phosphate mutase n=1 Tax=Py... 83 1e-14
UniRef100_C1G164 Phosphoacetylglucosamine mutase n=1 Tax=Paracoc... 83 1e-14
UniRef100_C0S3D9 Phosphoacetylglucosamine mutase n=1 Tax=Paracoc... 83 1e-14
UniRef100_Q6BSY8 DEHA2D04972p n=1 Tax=Debaryomyces hansenii RepI... 82 2e-14
UniRef100_C4V755 Putative uncharacterized protein n=1 Tax=Nosema... 82 2e-14
UniRef100_Q2UNB6 Phosphoglucomutase/phosphomannomutase n=1 Tax=A... 81 4e-14
UniRef100_Q1E1N4 Putative uncharacterized protein n=1 Tax=Coccid... 81 4e-14
UniRef100_C5P9Y4 Phosphoglucomutase/phosphomannomutase, putative... 81 4e-14
UniRef100_C5MC59 Phosphoacetylglucosamine mutase n=1 Tax=Candida... 81 4e-14
UniRef100_C5FDM1 N-acetylglucosamine-phosphate mutase n=1 Tax=Mi... 81 4e-14
UniRef100_B8NNC4 N-acetylglucosamine-phosphate mutase n=1 Tax=As... 81 4e-14
UniRef100_A5DNZ9 Putative uncharacterized protein n=1 Tax=Pichia... 81 5e-14
UniRef100_Q4WJF0 N-acetylglucosamine-phosphate mutase n=1 Tax=As... 80 7e-14
UniRef100_B0XPI4 N-acetylglucosamine-phosphate mutase n=1 Tax=As... 80 7e-14
UniRef100_A2RB18 Contig An18c0160, complete genome. (Fragment) n... 80 7e-14
UniRef100_A1D406 N-acetylglucosamine-phosphate mutase n=1 Tax=Ne... 80 7e-14
UniRef100_A1CR31 N-acetylglucosamine-phosphate mutase n=1 Tax=As... 80 7e-14
UniRef100_C4QF48 Phosphoglucomutase, putative n=1 Tax=Schistosom... 80 9e-14
UniRef100_Q5B5E6 Putative uncharacterized protein n=1 Tax=Emeric... 80 9e-14
UniRef100_C8V496 Predicted phosphoacetylglucosamine mutase (Euro... 80 9e-14
UniRef100_Q6FXI7 Strain CBS138 chromosome B complete sequence n=... 80 1e-13
UniRef100_Q5AKW4 Putative uncharacterized protein AGM1 n=1 Tax=C... 79 1e-13
UniRef100_C4YFV0 Phosphoacetylglucosamine mutase n=1 Tax=Candida... 79 1e-13
UniRef100_B9W9Z4 Phosphoacetylglucosamine mutase, putative (Acet... 79 1e-13
UniRef100_Q9P4V2 Phosphoacetylglucosamine mutase n=1 Tax=Candida... 79 1e-13
UniRef100_UPI0000F24296 Phosphoacetylglucosamine Mutase n=1 Tax=... 79 2e-13
UniRef100_Q0CTJ5 Putative uncharacterized protein n=1 Tax=Asperg... 79 2e-13
UniRef100_A8PZW5 Putative uncharacterized protein n=1 Tax=Malass... 79 2e-13
UniRef100_A3GI60 Phosphoacetylglucosamine Mutase n=1 Tax=Pichia ... 79 2e-13
UniRef100_Q75CE5 ACR015Wp n=1 Tax=Eremothecium gossypii RepID=Q7... 79 3e-13
UniRef100_Q0UJJ8 Putative uncharacterized protein n=1 Tax=Phaeos... 79 3e-13
UniRef100_Q5DF02 SJCHGC03861 protein n=1 Tax=Schistosoma japonic... 78 3e-13
UniRef100_C7TY12 Phosphoglucomutase 3 n=1 Tax=Schistosoma japoni... 78 3e-13
UniRef100_B6JX50 Phosphoacetylglucosamine mutase n=1 Tax=Schizos... 77 6e-13
UniRef100_A8PY91 Phosphoglucomutase/phosphomannomutase, C-termin... 77 7e-13
UniRef100_Q57XH7 Phosphoacetylglucosamine mutase, putative n=1 T... 77 1e-12
UniRef100_C9ZUL6 Putative uncharacterized protein n=1 Tax=Trypan... 77 1e-12
UniRef100_B6H1E1 Pc13g02740 protein n=1 Tax=Penicillium chrysoge... 76 1e-12
UniRef100_A4S8A5 Predicted protein n=1 Tax=Ostreococcus lucimari... 76 2e-12
UniRef100_A4RMS5 Putative uncharacterized protein n=1 Tax=Magnap... 76 2e-12
UniRef100_A4HTA8 Phosphoacetylglucosamine mutase-like protein (A... 75 2e-12
UniRef100_Q5KH62 Phosphoacetylglucosamine mutase, putative n=1 T... 75 2e-12
UniRef100_Q00UB2 Putative N-acetylglucosamine-phosphate mutase (... 75 3e-12
UniRef100_Q6C454 YALI0E29579p n=1 Tax=Yarrowia lipolytica RepID=... 75 3e-12
UniRef100_A5E1Q6 Phosphoacetylglucosamine mutase n=1 Tax=Loddero... 74 5e-12
UniRef100_Q4QIK7 Phosphoacetylglucosamine mutase-like protein (A... 74 6e-12
UniRef100_Q6CSH9 KLLA0D00858p n=1 Tax=Kluyveromyces lactis RepID... 74 6e-12
UniRef100_C7YKF4 Predicted protein n=1 Tax=Nectria haematococca ... 74 6e-12
UniRef100_A8XP38 Putative uncharacterized protein n=1 Tax=Caenor... 74 8e-12
UniRef100_Q8SSL7 Probable phosphoacetylglucosamine mutase n=1 Ta... 74 8e-12
UniRef100_UPI000023D61E hypothetical protein FG01133.1 n=1 Tax=G... 73 1e-11
UniRef100_Q4P4I0 Putative uncharacterized protein n=1 Tax=Ustila... 73 1e-11
UniRef100_Q7SD48 Putative uncharacterized protein n=1 Tax=Neuros... 72 2e-11
UniRef100_Q19680 Protein F21D5.1, confirmed by transcript eviden... 71 4e-11
UniRef100_C8Z6T4 Pcm1p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 70 9e-11
UniRef100_C7GXE5 Pcm1p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 70 9e-11
UniRef100_B5VH78 YEL058Wp-like protein n=1 Tax=Saccharomyces cer... 70 9e-11
UniRef100_B3LRW0 Phosphoacetylglucosamine mutase n=1 Tax=Sacchar... 70 9e-11
UniRef100_A6ZQP3 Phosphoacetylglucosamine mutase n=1 Tax=Sacchar... 70 9e-11
UniRef100_P38628 Phosphoacetylglucosamine mutase n=1 Tax=Sacchar... 70 9e-11
UniRef100_C4QW03 Essential N-acetylglucosamine-phosphate mutase ... 70 1e-10
UniRef100_A4H3S0 Phosphoacetylglucosamine mutase-like gene n=1 T... 69 2e-10
UniRef100_A7TGV8 Putative uncharacterized protein n=1 Tax=Vander... 69 2e-10
UniRef100_C5DWY0 ZYRO0F00506p n=1 Tax=Zygosaccharomyces rouxii C... 69 2e-10
UniRef100_UPI0001926A08 PREDICTED: similar to phosphoglucomutase... 68 3e-10
UniRef100_Q23DK4 Phosphoglucomutase/phosphomannomutase, C-termin... 64 6e-09
UniRef100_Q4DHI5 Phosphoacetylglucosamine mutase, putative n=1 T... 62 2e-08
UniRef100_Q4DA33 Phosphoacetylglucosamine mutase, putative n=1 T... 62 2e-08
UniRef100_Q2HFH7 Putative uncharacterized protein n=1 Tax=Chaeto... 62 2e-08
UniRef100_B6K2S5 Phosphoacetylglucosamine mutase n=1 Tax=Schizos... 62 3e-08
UniRef100_C4J5K8 Putative uncharacterized protein n=1 Tax=Zea ma... 57 8e-07
UniRef100_C0HHR6 Putative uncharacterized protein n=1 Tax=Zea ma... 57 8e-07
UniRef100_UPI0001925A99 PREDICTED: hypothetical protein, partial... 45 1e-06
UniRef100_Q1D3Z1 Response regulator n=1 Tax=Myxococcus xanthus D... 41 2e-06
UniRef100_C1E6Z7 Predicted protein n=1 Tax=Micromonas sp. RCC299... 55 4e-06
UniRef100_A9P3B7 Anther-specific proline rich protein n=1 Tax=Br... 44 7e-06
UniRef100_A9P3B4 Anther-specific proline rich protein n=1 Tax=Br... 44 7e-06
UniRef100_A9P3B0 Anther-specific proline rich protein n=1 Tax=Br... 44 7e-06
UniRef100_A9P3A9 Anther-specific proline rich protein n=4 Tax=Br... 44 7e-06
UniRef100_A9P3A8 Anther-specific proline rich protein n=1 Tax=Br... 44 7e-06
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 44 8e-06