[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AU193995 PFL053g06_r
(493 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
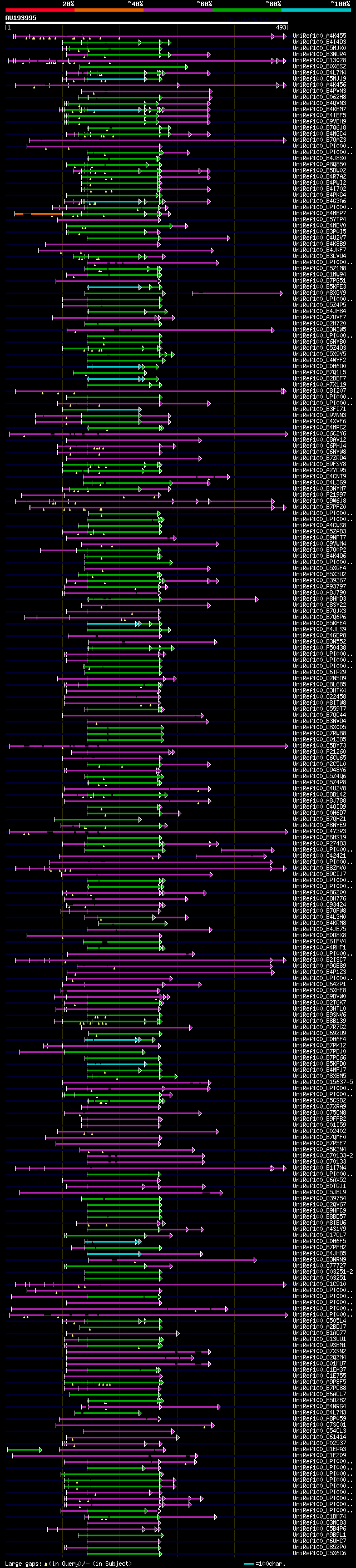
significant alignments:[graphical|details]
UniRef100_A4K455 Antifreeze glycoprotein n=1 Tax=Boreogadus said... 71 3e-11
UniRef100_B4I4D3 GM10561 n=1 Tax=Drosophila sechellia RepID=B4I4... 69 1e-10
UniRef100_C5MJK0 Predicted protein n=1 Tax=Candida tropicalis MY... 69 2e-10
UniRef100_B3NUR4 GG19208 n=1 Tax=Drosophila erecta RepID=B3NUR4_... 68 3e-10
UniRef100_O13028 Antifreeze glycopeptide AFGP polyprotein n=1 Ta... 68 4e-10
UniRef100_B0X8S2 Guanylate cyclase n=1 Tax=Culex quinquefasciatu... 67 5e-10
UniRef100_B4L7M4 GI11200 n=1 Tax=Drosophila mojavensis RepID=B4L... 67 6e-10
UniRef100_C5MJJ9 Predicted protein n=1 Tax=Candida tropicalis MY... 67 8e-10
UniRef100_A4K456 Antifreeze glycoprotein (Fragment) n=1 Tax=Bore... 66 1e-09
UniRef100_B4PVN3 GE24109 n=1 Tax=Drosophila yakuba RepID=B4PVN3_... 66 1e-09
UniRef100_Q062H8 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 66 1e-09
UniRef100_B4QVN3 GD19174 n=1 Tax=Drosophila simulans RepID=B4QVN... 66 1e-09
UniRef100_B4KBM7 GI23172 n=1 Tax=Drosophila mojavensis RepID=B4K... 66 1e-09
UniRef100_B4IBF5 GM15249 n=1 Tax=Drosophila sechellia RepID=B4IB... 66 1e-09
UniRef100_Q9VEH9 CG14327 n=1 Tax=Drosophila melanogaster RepID=Q... 65 2e-09
UniRef100_B7Q6J8 Secreted salivary gland peptide, putative n=1 T... 65 2e-09
UniRef100_B4MGC4 GJ18490 n=1 Tax=Drosophila virilis RepID=B4MGC4... 65 2e-09
UniRef100_B7QAZ3 Secreted salivary gland peptide, putative (Frag... 65 3e-09
UniRef100_UPI0001745B20 RNA-binding region RNP-1 n=1 Tax=Verruco... 65 3e-09
UniRef100_UPI0000DB6D77 PREDICTED: similar to CG5913-PA n=1 Tax=... 65 3e-09
UniRef100_B4J8S0 GH19937 n=1 Tax=Drosophila grimshawi RepID=B4J8... 65 3e-09
UniRef100_A8Q850 Putative uncharacterized protein n=1 Tax=Malass... 65 3e-09
UniRef100_B5DW02 GA27272 n=1 Tax=Drosophila pseudoobscura pseudo... 64 4e-09
UniRef100_B4R7A2 GD17424 n=1 Tax=Drosophila simulans RepID=B4R7A... 64 4e-09
UniRef100_B4PWI2 GE17772 n=1 Tax=Drosophila yakuba RepID=B4PWI2_... 64 4e-09
UniRef100_B4I702 GM22940 n=1 Tax=Drosophila sechellia RepID=B4I7... 64 4e-09
UniRef100_B4PKG4 GE25486 n=1 Tax=Drosophila yakuba RepID=B4PKG4_... 64 5e-09
UniRef100_B4G3A6 GL23478 n=1 Tax=Drosophila persimilis RepID=B4G... 64 5e-09
UniRef100_UPI0000DB6CCB PREDICTED: hypothetical protein n=1 Tax=... 64 7e-09
UniRef100_B4MBP7 GJ14488 n=1 Tax=Drosophila virilis RepID=B4MBP7... 63 9e-09
UniRef100_C5YTP4 Putative uncharacterized protein Sb08g006730 n=... 63 1e-08
UniRef100_B4MEV0 GJ14886 n=1 Tax=Drosophila virilis RepID=B4MEV0... 63 1e-08
UniRef100_B3P0I5 GG22373 n=1 Tax=Drosophila erecta RepID=B3P0I5_... 63 1e-08
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 62 2e-08
UniRef100_B4K8B9 GI22197 n=1 Tax=Drosophila mojavensis RepID=B4K... 62 2e-08
UniRef100_B4JKF7 GH12057 n=1 Tax=Drosophila grimshawi RepID=B4JK... 62 2e-08
UniRef100_B3LVU4 GF18623 n=1 Tax=Drosophila ananassae RepID=B3LV... 62 2e-08
UniRef100_UPI00015B637D PREDICTED: similar to conserved hypothet... 62 2e-08
UniRef100_C5Z1M8 Putative uncharacterized protein Sb10g012110 n=... 62 2e-08
UniRef100_Q1MW94 Shematrin-3 n=1 Tax=Pinctada fucata RepID=Q1MW9... 62 2e-08
UniRef100_B7PG51 Putative uncharacterized protein (Fragment) n=1... 62 2e-08
UniRef100_B5KFE3 KRMP-5 n=1 Tax=Pinctada margaritifera RepID=B5K... 62 2e-08
UniRef100_A8XGY9 C. briggsae CBR-RNH-1.1 protein n=1 Tax=Caenorh... 57 2e-08
UniRef100_UPI0000E127A2 Os06g0317400 n=1 Tax=Oryza sativa Japoni... 62 3e-08
UniRef100_Q5Z4P5 Os06g0317400 protein n=1 Tax=Oryza sativa Japon... 62 3e-08
UniRef100_B4JH84 GH19524 n=1 Tax=Drosophila grimshawi RepID=B4JH... 62 3e-08
UniRef100_A7UVF7 AGAP011901-PA (Fragment) n=1 Tax=Anopheles gamb... 62 3e-08
UniRef100_Q2H720 ATP-dependent RNA helicase DBP2 n=1 Tax=Chaetom... 62 3e-08
UniRef100_B3N3W5 GG23771 n=1 Tax=Drosophila erecta RepID=B3N3W5_... 61 3e-08
UniRef100_UPI00004367E2 hypothetical protein LOC323529 n=1 Tax=D... 61 3e-08
UniRef100_Q6NYB0 Zgc:77366 n=1 Tax=Danio rerio RepID=Q6NYB0_DANRE 61 3e-08
UniRef100_Q5Z4Q3 Os06g0316800 protein n=1 Tax=Oryza sativa Japon... 61 3e-08
UniRef100_C5X9Y5 Putative uncharacterized protein Sb02g034435 n=... 61 3e-08
UniRef100_C4WYF2 ACYPI004778 protein n=1 Tax=Acyrthosiphon pisum... 61 3e-08
UniRef100_C0H6D0 Putative cuticle protein n=1 Tax=Bombyx mori Re... 61 3e-08
UniRef100_B7Q1L5 Secreted protein, putative (Fragment) n=1 Tax=I... 61 3e-08
UniRef100_B2DBF7 Cuticular protein CPG12 n=1 Tax=Papilio xuthus ... 61 3e-08
UniRef100_A7X119 KRMP-9 n=1 Tax=Pinctada margaritifera RepID=A7X... 61 3e-08
UniRef100_Q8I207 Putative uncharacterized protein n=1 Tax=Plasmo... 61 4e-08
UniRef100_UPI0000DB6C07 PREDICTED: hypothetical protein n=1 Tax=... 61 4e-08
UniRef100_UPI0001A2D182 heterogeneous nuclear ribonucleoprotein ... 61 4e-08
UniRef100_B3FI71 Prion protein n=1 Tax=Poecilia reticulata RepID... 61 4e-08
UniRef100_Q9VNN3 CG15597 n=1 Tax=Drosophila melanogaster RepID=Q... 61 4e-08
UniRef100_C4XVF6 MIP10548p (Fragment) n=1 Tax=Drosophila melanog... 61 4e-08
UniRef100_B4MPC2 GK21632 n=1 Tax=Drosophila willistoni RepID=B4M... 61 4e-08
UniRef100_Q6C2Y6 Metacaspase-1 n=1 Tax=Yarrowia lipolytica RepID... 61 4e-08
UniRef100_Q8AV12 Adult keratin XAK-C n=1 Tax=Xenopus laevis RepI... 60 6e-08
UniRef100_Q6PHJ4 Hnrpa0 protein (Fragment) n=1 Tax=Danio rerio R... 60 6e-08
UniRef100_Q6NYW8 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 60 6e-08
UniRef100_B7ZRD4 Ak-c-A protein n=1 Tax=Xenopus laevis RepID=B7Z... 60 6e-08
UniRef100_B9FSY8 Putative uncharacterized protein n=1 Tax=Oryza ... 60 6e-08
UniRef100_A2YC95 Putative uncharacterized protein n=1 Tax=Oryza ... 60 6e-08
UniRef100_Q4CNT9 Dispersed gene family protein 1 (DGF-1), putati... 60 6e-08
UniRef100_B4L3G9 GI15544 n=1 Tax=Drosophila mojavensis RepID=B4L... 60 6e-08
UniRef100_B3NYM7 GG10533 n=1 Tax=Drosophila erecta RepID=B3NYM7_... 60 6e-08
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 60 6e-08
UniRef100_Q9W6J8 Chimeric AFGP/trypsinogen-like serine protease ... 60 7e-08
UniRef100_B7PFZ0 Proteophosphoglycan, putative (Fragment) n=1 Ta... 60 7e-08
UniRef100_UPI00016E9D6C UPI00016E9D6C related cluster n=1 Tax=Ta... 60 8e-08
UniRef100_UPI00016E7C99 UPI00016E7C99 related cluster n=1 Tax=Ta... 60 8e-08
UniRef100_A4CWS8 RNA-binding region RNP-1 (RNA recognition motif... 60 8e-08
UniRef100_Q5ZAB3 Os06g0317600 protein n=1 Tax=Oryza sativa Japon... 60 8e-08
UniRef100_B9NFT7 Predicted protein n=1 Tax=Populus trichocarpa R... 60 8e-08
UniRef100_Q9VWM4 CG14191 n=1 Tax=Drosophila melanogaster RepID=Q... 60 8e-08
UniRef100_B7Q0P2 Putative uncharacterized protein (Fragment) n=1... 60 8e-08
UniRef100_B4K4Q6 GI23003 n=1 Tax=Drosophila mojavensis RepID=B4K... 60 8e-08
UniRef100_UPI00005A1BE2 PREDICTED: similar to keratin 13 isoform... 60 1e-07
UniRef100_Q5XGF4 Heterogeneous nuclear ribonucleoprotein A1 n=1 ... 60 1e-07
UniRef100_B5X3U2 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 60 1e-07
UniRef100_Q39367 Glycine-rich protein (Fragment) n=1 Tax=Brassic... 60 1e-07
UniRef100_P93797 Pherophorin-S n=1 Tax=Volvox carteri RepID=P937... 60 1e-07
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 60 1e-07
UniRef100_A8HMD3 Putative uncharacterized protein n=1 Tax=Chlamy... 60 1e-07
UniRef100_Q8SY22 RE09269p (Fragment) n=1 Tax=Drosophila melanoga... 60 1e-07
UniRef100_B7QJX3 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_B7Q6P6 Putative uncharacterized protein (Fragment) n=1... 60 1e-07
UniRef100_B5KFE4 KRMP-6 n=1 Tax=Pinctada margaritifera RepID=B5K... 60 1e-07
UniRef100_B4JLS9 GH24514 n=1 Tax=Drosophila grimshawi RepID=B4JL... 60 1e-07
UniRef100_B4GDP8 GL21516 n=1 Tax=Drosophila persimilis RepID=B4G... 60 1e-07
UniRef100_B3N552 GG10371 n=1 Tax=Drosophila erecta RepID=B3N552_... 60 1e-07
UniRef100_P50438 Uncharacterized protein F12A10.7 n=1 Tax=Caenor... 60 1e-07
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 59 1e-07
UniRef100_UPI000155382B PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_UPI0000DB7202 PREDICTED: hypothetical protein n=1 Tax=... 59 1e-07
UniRef100_Q6IP29 LOC397751 protein n=1 Tax=Xenopus laevis RepID=... 59 1e-07
UniRef100_Q2N5D9 Autotransporter n=1 Tax=Erythrobacter litoralis... 59 1e-07
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 59 1e-07
UniRef100_Q3HTK4 Cell wall protein pherophorin-C3 n=1 Tax=Chlamy... 59 1e-07
UniRef100_O22458 Hydroxyproline-rich glycoprotein gas28p n=1 Tax... 59 1e-07
UniRef100_A8ITW8 Dicer-like protein n=1 Tax=Chlamydomonas reinha... 59 1e-07
UniRef100_Q559T7 Putative uncharacterized protein n=1 Tax=Dictyo... 59 1e-07
UniRef100_B7QC44 Proline-rich protein, putative (Fragment) n=1 T... 59 1e-07
UniRef100_B3NVD4 GG18375 n=1 Tax=Drosophila erecta RepID=B3NVD4_... 59 1e-07
UniRef100_Q8X005 Glycine rich protein (Het-COR) n=1 Tax=Neurospo... 59 1e-07
UniRef100_Q7RW88 Putative uncharacterized protein n=1 Tax=Neuros... 59 1e-07
UniRef100_Q01385 Glycine rich protein (Fragment) n=1 Tax=Neurosp... 59 1e-07
UniRef100_C5DY73 ZYRO0F10802p n=1 Tax=Zygosaccharomyces rouxii C... 59 1e-07
UniRef100_P21260 Uncharacterized proline-rich protein (Fragment)... 59 1e-07
UniRef100_C6CW65 Putative uncharacterized protein n=1 Tax=Paenib... 59 2e-07
UniRef100_A2C5L0 RNA-binding region RNP-1 (RNA recognition motif... 59 2e-07
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 59 2e-07
UniRef100_Q5Z4Q6 Os06g0316300 protein n=1 Tax=Oryza sativa Japon... 59 2e-07
UniRef100_Q5Z4P8 cDNA clone:001-103-C11, full insert sequence n=... 59 2e-07
UniRef100_Q4U2V8 Hydroxyproline-rich glycoprotein GAS28 n=1 Tax=... 59 2e-07
UniRef100_B8B142 Putative uncharacterized protein n=1 Tax=Oryza ... 59 2e-07
UniRef100_A8J788 Hydroxyproline-rich glycoprotein, stress-induce... 59 2e-07
UniRef100_Q4QIQ9 ATP-dependent DEAD/H RNA helicase, putative n=1... 59 2e-07
UniRef100_C0H6D7 Putative cuticle protein n=1 Tax=Bombyx mori Re... 59 2e-07
UniRef100_B7QHZ1 Cement protein, putative n=1 Tax=Ixodes scapula... 59 2e-07
UniRef100_A8NYE9 Putative uncharacterized protein n=1 Tax=Brugia... 59 2e-07
UniRef100_C4Y3R3 Putative uncharacterized protein n=1 Tax=Clavis... 59 2e-07
UniRef100_B6HS19 Pc22g21030 protein n=1 Tax=Penicillium chrysoge... 59 2e-07
UniRef100_P27483 Glycine-rich cell wall structural protein n=1 T... 59 2e-07
UniRef100_UPI0000123EDF Hypothetical protein CBG13051 n=1 Tax=Ca... 57 2e-07
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 50 2e-07
UniRef100_UPI000069F113 Myeloid/lymphoid or mixed-lineage leukem... 59 2e-07
UniRef100_B8ZMV0 Cell wall surface anchored protein n=1 Tax=Stre... 59 2e-07
UniRef100_B9CIJ7 Chemotaxis protein CheA n=1 Tax=Burkholderia mu... 59 2e-07
UniRef100_UPI000150401A hypothetical protein MGG_07033 n=1 Tax=M... 59 2e-07
UniRef100_UPI0000E498A8 PREDICTED: hypothetical protein, partial... 59 2e-07
UniRef100_A8G200 RNA-binding region RNP-1 (RNA recognition motif... 59 2e-07
UniRef100_Q8H776 Putative uncharacterized protein n=1 Tax=Arabid... 59 2e-07
UniRef100_Q93424 Protein E02A10.2, partially confirmed by transc... 59 2e-07
UniRef100_B7QFW8 Putative uncharacterized protein (Fragment) n=1... 59 2e-07
UniRef100_B4L3H0 GI15545 n=1 Tax=Drosophila mojavensis RepID=B4L... 59 2e-07
UniRef100_B4KRM8 GI19894 n=1 Tax=Drosophila mojavensis RepID=B4K... 59 2e-07
UniRef100_B4JE75 GH11326 n=1 Tax=Drosophila grimshawi RepID=B4JE... 59 2e-07
UniRef100_B0D8X8 Predicted protein n=1 Tax=Laccaria bicolor S238... 59 2e-07
UniRef100_Q6IFV4 Keratin, type I cytoskeletal 13 n=1 Tax=Rattus ... 59 2e-07
UniRef100_A4RHF1 ATP-dependent RNA helicase DED1 n=1 Tax=Magnapo... 59 2e-07
UniRef100_UPI00015B62B9 PREDICTED: similar to conserved hypothet... 58 3e-07
UniRef100_B2ISC7 Cell wall surface anchor family protein n=1 Tax... 58 3e-07
UniRef100_A9GE89 Putative uncharacterized protein n=1 Tax=Sorang... 58 3e-07
UniRef100_B4P1Z3 GE18575 n=1 Tax=Drosophila yakuba RepID=B4P1Z3_... 58 3e-07
UniRef100_UPI0001796C05 PREDICTED: similar to LOC507464 protein ... 58 3e-07
UniRef100_Q642P1 LOC446967 protein (Fragment) n=1 Tax=Xenopus la... 58 3e-07
UniRef100_Q5XHE8 Putative uncharacterized protein n=1 Tax=Xenopu... 58 3e-07
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 58 3e-07
UniRef100_B2T6K7 Putative lipoprotein n=1 Tax=Burkholderia phyto... 58 3e-07
UniRef100_Q3HTL0 Pherophorin-V1 protein n=1 Tax=Volvox carteri f... 58 3e-07
UniRef100_B9SNV6 Glycine-rich cell wall structural protein 1.8, ... 58 3e-07
UniRef100_B8B139 Putative uncharacterized protein n=1 Tax=Oryza ... 58 3e-07
UniRef100_A7R7G2 Chromosome undetermined scaffold_1755, whole ge... 58 3e-07
UniRef100_Q692U9 Cement protein n=1 Tax=Ixodes scapularis RepID=... 58 3e-07
UniRef100_C0H6F4 Putative cuticle protein n=1 Tax=Bombyx mori Re... 58 3e-07
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 58 3e-07
UniRef100_B7PDJ0 Submaxillary gland androgen-regulated protein 3... 58 3e-07
UniRef100_B7PC66 Putative uncharacterized protein (Fragment) n=1... 58 3e-07
UniRef100_B5KFD0 Shematrin-8 n=1 Tax=Pinctada margaritifera RepI... 58 3e-07
UniRef100_B4MFJ7 GJ15063 n=1 Tax=Drosophila virilis RepID=B4MFJ7... 58 3e-07
UniRef100_A8XBM5 C. briggsae CBR-COL-103 protein n=1 Tax=Caenorh... 58 3e-07
UniRef100_Q15637-5 Isoform 5 of Splicing factor 1 n=1 Tax=Homo s... 58 3e-07
UniRef100_UPI0001A7B233 nucleic acid binding n=1 Tax=Arabidopsis... 58 4e-07
UniRef100_UPI000155F1A7 PREDICTED: similar to keratin 13 n=1 Tax... 58 4e-07
UniRef100_C5CSB2 RNP-1 like RNA-binding protein n=1 Tax=Variovor... 58 4e-07
UniRef100_Q7XRA9 Os04g0438101 protein n=1 Tax=Oryza sativa Japon... 58 4e-07
UniRef100_Q75QN8 Cold shock domain protein 3 n=1 Tax=Triticum ae... 58 4e-07
UniRef100_B9FFB2 Putative uncharacterized protein n=1 Tax=Oryza ... 58 4e-07
UniRef100_Q01I59 H0315A08.9 protein n=2 Tax=Oryza sativa RepID=Q... 58 4e-07
UniRef100_O02402 Insoluble protein n=1 Tax=Pinctada fucata RepID... 58 4e-07
UniRef100_B7QMF0 Putative uncharacterized protein (Fragment) n=1... 58 4e-07
UniRef100_B7P5E7 Putative uncharacterized protein (Fragment) n=1... 58 4e-07
UniRef100_A5K3N4 Putative uncharacterized protein n=1 Tax=Plasmo... 58 4e-07
UniRef100_O70133-2 Isoform 2 of ATP-dependent RNA helicase A n=1... 58 4e-07
UniRef100_O70133 ATP-dependent RNA helicase A n=2 Tax=Mus muscul... 58 4e-07
UniRef100_B1I7N4 Cell wall surface anchor family protein n=1 Tax... 57 5e-07
UniRef100_UPI0001923E9F PREDICTED: similar to nematocyst outer w... 57 5e-07
UniRef100_Q6AX52 LOC397907 protein (Fragment) n=1 Tax=Xenopus la... 57 5e-07
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 57 5e-07
UniRef100_C5JBL9 Uncharacterized protein n=1 Tax=uncultured bact... 57 5e-07
UniRef100_Q39754 GRPF1 n=1 Tax=Fagus sylvatica RepID=Q39754_FAGSY 57 5e-07
UniRef100_Q2QV67 Expressed protein n=1 Tax=Oryza sativa Japonica... 57 5e-07
UniRef100_B9HFC9 Predicted protein n=1 Tax=Populus trichocarpa R... 57 5e-07
UniRef100_B8BD57 Putative uncharacterized protein n=1 Tax=Oryza ... 57 5e-07
UniRef100_A8IBU6 Nuclear ribonucleoprotein n=1 Tax=Chlamydomonas... 57 5e-07
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 57 5e-07
UniRef100_Q17QL7 KRT15 protein n=1 Tax=Bos taurus RepID=Q17QL7_B... 57 5e-07
UniRef100_C0H6F5 Putative cuticle protein n=1 Tax=Bombyx mori Re... 57 5e-07
UniRef100_B7PFH2 Secreted salivary gland peptide, putative (Frag... 57 5e-07
UniRef100_B4JH85 GH18097 n=1 Tax=Drosophila grimshawi RepID=B4JH... 57 5e-07
UniRef100_B3NRN9 GG22510 n=1 Tax=Drosophila erecta RepID=B3NRN9_... 57 5e-07
UniRef100_O77727 Keratin, type I cytoskeletal 15 n=1 Tax=Ovis ar... 57 5e-07
UniRef100_Q03251-2 Isoform 2 of Glycine-rich RNA-binding protein... 57 5e-07
UniRef100_Q03251 Glycine-rich RNA-binding protein 8 n=2 Tax=Arab... 57 5e-07
UniRef100_C1C910 Cell wall surface anchor family protein n=1 Tax... 57 6e-07
UniRef100_UPI00019847F3 PREDICTED: hypothetical protein n=1 Tax=... 57 6e-07
UniRef100_UPI0001982DE4 PREDICTED: similar to leucine-rich repea... 57 6e-07
UniRef100_UPI0000F2E52D PREDICTED: similar to frizzled homolog 8... 57 6e-07
UniRef100_UPI0000E11F6B Os03g0190000 n=1 Tax=Oryza sativa Japoni... 57 6e-07
UniRef100_UPI00003BE83A hypothetical protein DEHA0G23474g n=1 Ta... 57 6e-07
UniRef100_Q505L4 LOC397864 protein n=1 Tax=Xenopus laevis RepID=... 57 6e-07
UniRef100_A2BDJ7 Prion protein-like protein PrP-like n=1 Tax=Gas... 57 6e-07
UniRef100_B1AQ77 Keratin 15 n=1 Tax=Mus musculus RepID=B1AQ77_MOUSE 57 6e-07
UniRef100_Q13UU1 Putative lipoprotein n=1 Tax=Burkholderia xenov... 57 6e-07
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 57 6e-07
UniRef100_Q7XSN2 OSJNBb0028M18.5 protein n=1 Tax=Oryza sativa Ja... 57 6e-07
UniRef100_Q2QZM4 Transposon protein, putative, CACTA, En/Spm sub... 57 6e-07
UniRef100_Q01MU7 OSIGBa0102O13.9 protein n=1 Tax=Oryza sativa Re... 57 6e-07
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 57 6e-07
UniRef100_C1E755 Putative uncharacterized protein n=1 Tax=Microm... 57 6e-07
UniRef100_A9P8F5 Putative uncharacterized protein n=1 Tax=Populu... 57 6e-07
UniRef100_B7PC88 Protein tonB, putative (Fragment) n=1 Tax=Ixode... 57 6e-07
UniRef100_B6ACL7 Putative uncharacterized protein n=1 Tax=Crypto... 57 6e-07
UniRef100_B5DZB2 GA24506 n=1 Tax=Drosophila pseudoobscura pseudo... 57 6e-07
UniRef100_B4NRG4 GK17702 (Fragment) n=1 Tax=Drosophila williston... 57 6e-07
UniRef100_B4L7M3 GI11199 n=1 Tax=Drosophila mojavensis RepID=B4L... 57 6e-07
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 57 6e-07
UniRef100_Q7SC01 Putative uncharacterized protein n=1 Tax=Neuros... 57 6e-07
UniRef100_Q54CL3 Probable peroxisomal membrane protein PEX13 n=1... 57 6e-07
UniRef100_Q61414 Keratin, type I cytoskeletal 15 n=1 Tax=Mus mus... 57 6e-07
UniRef100_P02537 Keratin-3, type I cytoskeletal 51 kDa n=1 Tax=X... 57 6e-07
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 54 7e-07
UniRef100_C1E209 KH domain-containing protein n=1 Tax=Micromonas... 57 8e-07
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 57 8e-07
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 57 8e-07
UniRef100_UPI000155434A PREDICTED: similar to olfactory receptor... 57 8e-07
UniRef100_UPI0000E4FB5A keratin 13 isoform a n=1 Tax=Homo sapien... 57 8e-07
UniRef100_UPI0000E4FB59 keratin 13 isoform b n=1 Tax=Homo sapien... 57 8e-07
UniRef100_UPI0000DB7892 PREDICTED: similar to Osiris 16 CG31561-... 57 8e-07
UniRef100_UPI0000DA1F7B PREDICTED: similar to Loricrin n=1 Tax=R... 57 8e-07
UniRef100_UPI0000DC09EF UPI0000DC09EF related cluster n=1 Tax=Ra... 57 8e-07
UniRef100_UPI0000ECD10C Zinc finger homeobox protein 4 (Zinc fin... 57 8e-07
UniRef100_C1BM74 Heterogeneous nuclear ribonucleoprotein A0 n=1 ... 57 8e-07
UniRef100_Q3MC83 Putative uncharacterized protein n=1 Tax=Anabae... 57 8e-07
UniRef100_C5B4P6 Putative uncharacterized protein n=1 Tax=Methyl... 57 8e-07
UniRef100_A9B9L1 RNA-binding region RNP-1 (RNA recognition motif... 57 8e-07
UniRef100_A6UHC7 Outer membrane autotransporter barrel domain n=... 57 8e-07
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 57 8e-07
UniRef100_C5X6C6 Putative uncharacterized protein Sb02g043650 n=... 57 8e-07
UniRef100_C5WUU1 Putative uncharacterized protein Sb01g044570 n=... 57 8e-07
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 57 8e-07
UniRef100_A2YLR5 Putative uncharacterized protein n=1 Tax=Oryza ... 57 8e-07
UniRef100_A2YC94 Putative uncharacterized protein n=1 Tax=Oryza ... 57 8e-07
UniRef100_Q7QDL5 AGAP003388-PA n=1 Tax=Anopheles gambiae RepID=Q... 57 8e-07
UniRef100_Q4Q5P5 ATP-dependent RNA helicase, putative n=1 Tax=Le... 57 8e-07
UniRef100_O45114 Collagen protein 103, confirmed by transcript e... 57 8e-07
UniRef100_B7Q7P5 Submaxillary gland androgen-regulated protein 3... 57 8e-07
UniRef100_B7Q3U3 Putative uncharacterized protein (Fragment) n=1... 57 8e-07
UniRef100_B4IZJ4 GH14508 n=1 Tax=Drosophila grimshawi RepID=B4IZ... 57 8e-07
UniRef100_A4I7K4 ATP-dependent RNA helicase, putative n=1 Tax=Le... 57 8e-07
UniRef100_A2DI20 Putative uncharacterized protein n=1 Tax=Tricho... 57 8e-07
UniRef100_C9JS29 Putative uncharacterized protein TNRC18 n=1 Tax... 57 8e-07
UniRef100_A8K2H9 cDNA FLJ78503, highly similar to Homo sapiens k... 57 8e-07
UniRef100_A1A4E9 Keratin 13 n=1 Tax=Homo sapiens RepID=A1A4E9_HUMAN 57 8e-07
UniRef100_B2AKS1 Translation initiation factor IF-2 n=1 Tax=Podo... 57 8e-07
UniRef100_O15417-2 Isoform 2 of Trinucleotide repeat-containing ... 57 8e-07
UniRef100_P18165 Loricrin n=1 Tax=Mus musculus RepID=LORI_MOUSE 57 8e-07
UniRef100_P13646-3 Isoform 3 of Keratin, type I cytoskeletal 13 ... 57 8e-07
UniRef100_P13646 Keratin, type I cytoskeletal 13 n=1 Tax=Homo sa... 57 8e-07
UniRef100_P10496 Glycine-rich cell wall structural protein 1.8 n... 57 8e-07
UniRef100_Q1AMQ0 Antifreeze glycoprotein n=1 Tax=Eleginus gracil... 56 1e-06
UniRef100_Q97P71 Cell wall surface anchor family protein n=1 Tax... 56 1e-06
UniRef100_UPI00005A1BE7 PREDICTED: similar to keratin 13 isoform... 56 1e-06
UniRef100_UPI00005A1BE6 PREDICTED: similar to keratin 13 isoform... 56 1e-06
UniRef100_UPI00005A1BE5 PREDICTED: similar to keratin 13 isoform... 56 1e-06
UniRef100_UPI0000220B01 Hypothetical protein CBG11408 n=1 Tax=Ca... 56 1e-06
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 56 1e-06
UniRef100_B4AM45 Putative uncharacterized protein n=1 Tax=Bacill... 56 1e-06
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 56 1e-06
UniRef100_B9MZR3 Predicted protein n=1 Tax=Populus trichocarpa R... 56 1e-06
UniRef100_B9HX27 Predicted protein n=1 Tax=Populus trichocarpa R... 56 1e-06
UniRef100_O61948 Ground-like (Grd related) protein 29 n=1 Tax=Ca... 56 1e-06
UniRef100_B7QA44 Putative uncharacterized protein n=1 Tax=Ixodes... 56 1e-06
UniRef100_B6KME4 RRM domain-containing protein n=1 Tax=Toxoplasm... 56 1e-06
UniRef100_B5KFE2 KRMP-4 n=1 Tax=Pinctada margaritifera RepID=B5K... 56 1e-06
UniRef100_B4NHC9 GK14205 n=1 Tax=Drosophila willistoni RepID=B4N... 56 1e-06
UniRef100_B4NET8 GK25701 n=1 Tax=Drosophila willistoni RepID=B4N... 56 1e-06
UniRef100_B4MWP9 GK15516 n=1 Tax=Drosophila willistoni RepID=B4M... 56 1e-06
UniRef100_B4JMZ8 GH24721 n=1 Tax=Drosophila grimshawi RepID=B4JM... 56 1e-06
UniRef100_B4HQ31 GM20296 n=1 Tax=Drosophila sechellia RepID=B4HQ... 56 1e-06
UniRef100_B4GDJ8 GL10844 n=1 Tax=Drosophila persimilis RepID=B4G... 56 1e-06
UniRef100_B3MYN1 GF22178 n=1 Tax=Drosophila ananassae RepID=B3MY... 56 1e-06
UniRef100_A8XD35 Putative uncharacterized protein n=1 Tax=Caenor... 56 1e-06
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 56 1e-06
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 55 1e-06
UniRef100_UPI00000216EF glyceraldehyde-3-phosphate dehydrogenase... 55 1e-06
UniRef100_UPI0000DA2F98 PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI00005A5E79 PREDICTED: similar to angiomotin isoform... 56 1e-06
UniRef100_B0TGZ6 Putative uncharacterized protein n=1 Tax=Heliob... 56 1e-06
UniRef100_C5KK36 Putative uncharacterized protein n=1 Tax=Perkin... 56 1e-06
UniRef100_B4NME2 GK23126 n=1 Tax=Drosophila willistoni RepID=B4N... 56 1e-06
UniRef100_B4MTP0 GK23793 n=1 Tax=Drosophila willistoni RepID=B4M... 56 1e-06
UniRef100_UPI00019847F4 PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI00016C4475 putative RNA-binding protein n=1 Tax=Gem... 56 1e-06
UniRef100_UPI00015FF5B0 piccolo isoform 2 n=1 Tax=Mus musculus R... 56 1e-06
UniRef100_UPI00015FA08D piccolo isoform 1 n=1 Tax=Mus musculus R... 56 1e-06
UniRef100_UPI0001555060 PREDICTED: similar to keratin 13 n=1 Tax... 56 1e-06
UniRef100_UPI000155453A PREDICTED: similar to keratin 14 (epider... 56 1e-06
UniRef100_UPI0000D57944 PREDICTED: hypothetical protein n=1 Tax=... 56 1e-06
UniRef100_UPI000069F4F9 Heterogeneous nuclear ribonucleoprotein ... 56 1e-06
UniRef100_UPI00016D3858 piccolo (presynaptic cytomatrix protein)... 56 1e-06
UniRef100_UPI00016D3827 piccolo (presynaptic cytomatrix protein)... 56 1e-06
UniRef100_UPI00015DF6C1 piccolo (presynaptic cytomatrix protein)... 56 1e-06
UniRef100_UPI00016E9D6B UPI00016E9D6B related cluster n=1 Tax=Ta... 56 1e-06
UniRef100_Q7SXQ3 Zgc:66127 n=1 Tax=Danio rerio RepID=Q7SXQ3_DANRE 56 1e-06
UniRef100_Q4SQV2 Chromosome 1 SCAF14529, whole genome shotgun se... 56 1e-06
UniRef100_Q4RBG5 Chromosome undetermined SCAF21509, whole genome... 56 1e-06
UniRef100_Q7V9D6 RNA-binding region RNP-1 (RNA recognition motif... 56 1e-06
UniRef100_Q0BCI9 Putative uncharacterized protein n=1 Tax=Burkho... 56 1e-06
UniRef100_Q10FE5 Retrotransposon protein, putative, Ty1-copia su... 56 1e-06
UniRef100_C1MIF9 Predicted protein n=1 Tax=Micromonas pusilla CC... 56 1e-06
UniRef100_C1E9S3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 56 1e-06
UniRef100_B9SFQ6 Glycine-rich cell wall structural protein 1, pu... 56 1e-06
UniRef100_B9MZQ9 Predicted protein n=1 Tax=Populus trichocarpa R... 56 1e-06
UniRef100_B8Q691 Glycine-rich cell wall protein n=1 Tax=Oryza gr... 56 1e-06
UniRef100_B6VA25 Putative glycine-rich RNA-binding protein n=1 T... 56 1e-06
UniRef100_A9SKS2 Predicted protein n=1 Tax=Physcomitrella patens... 56 1e-06
UniRef100_B3MW52 GF22323 n=1 Tax=Drosophila ananassae RepID=B3MW... 56 1e-06
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 56 1e-06
UniRef100_C7Z9J0 Predicted protein n=1 Tax=Nectria haematococca ... 56 1e-06
UniRef100_B3LJM7 Putative uncharacterized protein n=1 Tax=Saccha... 56 1e-06
UniRef100_Q9QYX7-2 Isoform 2 of Protein piccolo n=1 Tax=Mus musc... 56 1e-06
UniRef100_Q9QYX7-3 Isoform 3 of Protein piccolo n=1 Tax=Mus musc... 56 1e-06
UniRef100_Q9QYX7-4 Isoform 4 of Protein piccolo n=1 Tax=Mus musc... 56 1e-06
UniRef100_Q9QYX7 Protein piccolo n=1 Tax=Mus musculus RepID=PCLO... 56 1e-06
UniRef100_A3C5A7 Glycine-rich cell wall structural protein 2 n=2... 56 1e-06
UniRef100_Q5B0J9 ATP-dependent RNA helicase dbp2 n=2 Tax=Emerice... 56 1e-06
UniRef100_P05685 Chorion class B protein B.L1 (Fragment) n=1 Tax... 56 1e-06
UniRef100_P08915 Chorion class B protein M3A5 (Fragment) n=1 Tax... 56 1e-06
UniRef100_P08827 Chorion class B protein L11 n=1 Tax=Bombyx mori... 56 1e-06
UniRef100_UPI0001553800 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_B4M321 GJ19126 n=1 Tax=Drosophila virilis RepID=B4M321... 55 2e-06
UniRef100_B4K868 GI22794 n=1 Tax=Drosophila mojavensis RepID=B4K... 55 2e-06
UniRef100_UPI00019856A3 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI00017C3E59 PREDICTED: similar to Keratin 10 (epider... 55 2e-06
UniRef100_UPI0000F2BD8B PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI00005A1BE4 PREDICTED: similar to keratin 13 isoform... 55 2e-06
UniRef100_UPI000069F105 Formin-like protein 3 (Formin homology 2... 55 2e-06
UniRef100_UPI00016E9D6D UPI00016E9D6D related cluster n=1 Tax=Ta... 55 2e-06
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 55 2e-06
UniRef100_Q7ZX33 Hnrpa1 protein n=1 Tax=Xenopus laevis RepID=Q7Z... 55 2e-06
UniRef100_A5VB72 Filamentous haemagglutinin family outer membran... 55 2e-06
UniRef100_A1TWH4 RNP-1-like RNA-binding protein n=1 Tax=Acidovor... 55 2e-06
UniRef100_Q9M3Y2 Glycine-rich protein n=1 Tax=Triticum aestivum ... 55 2e-06
UniRef100_Q39337 Glycine-rich_protein_(Aa1-291) n=1 Tax=Brassica... 55 2e-06
UniRef100_Q00S27 Chromosome 19 contig 1, DNA sequence n=1 Tax=Os... 55 2e-06
UniRef100_B8B6P8 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_A8HQ72 SR protein factor n=1 Tax=Chlamydomonas reinhar... 55 2e-06
UniRef100_A4S5W2 Predicted protein n=1 Tax=Ostreococcus lucimari... 55 2e-06
UniRef100_A4RWC6 Predicted protein n=1 Tax=Ostreococcus lucimari... 55 2e-06
UniRef100_A6QNZ7 Keratin 10 (Epidermolytic hyperkeratosis; kerat... 55 2e-06
UniRef100_C0H6F3 Putative cuticle protein n=1 Tax=Bombyx mori Re... 55 2e-06
UniRef100_B7QF53 Putative uncharacterized protein n=1 Tax=Ixodes... 55 2e-06
UniRef100_B7Q768 Secreted protein, putative (Fragment) n=1 Tax=I... 55 2e-06
UniRef100_B5KFD1 Shematrin-9 n=1 Tax=Pinctada margaritifera RepI... 55 2e-06
UniRef100_B4QE04 GD25777 n=1 Tax=Drosophila simulans RepID=B4QE0... 55 2e-06
UniRef100_B4Q1P9 GE16203 n=1 Tax=Drosophila yakuba RepID=B4Q1P9_... 55 2e-06
UniRef100_B4PRW4 GE23709 n=1 Tax=Drosophila yakuba RepID=B4PRW4_... 55 2e-06
UniRef100_B4M120 GJ23081 n=1 Tax=Drosophila virilis RepID=B4M120... 55 2e-06
UniRef100_B4JKF6 GH12058 n=1 Tax=Drosophila grimshawi RepID=B4JK... 55 2e-06
UniRef100_B1Q4V4 Shematrin (Fragment) n=1 Tax=Pinctada maxima Re... 55 2e-06
UniRef100_A8X388 C. briggsae CBR-GRL-25 protein n=1 Tax=Caenorha... 55 2e-06
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 55 2e-06
UniRef100_Q08601 Metacaspase-1 n=5 Tax=Saccharomyces cerevisiae ... 55 2e-06
UniRef100_P06394 Keratin, type I cytoskeletal 10 n=1 Tax=Bos tau... 55 2e-06
UniRef100_Q64467 Glyceraldehyde-3-phosphate dehydrogenase, testi... 55 2e-06
UniRef100_B9PK59 Putative uncharacterized protein n=1 Tax=Toxopl... 55 2e-06
UniRef100_UPI0001796C4A PREDICTED: similar to keratin 10 n=1 Tax... 55 2e-06
UniRef100_UPI0001758286 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI0001556277 PREDICTED: hypothetical protein n=1 Tax=... 55 2e-06
UniRef100_UPI0000E1F767 PREDICTED: formin-like 2 isoform 3 n=1 T... 55 2e-06
UniRef100_UPI0000E1F766 PREDICTED: formin-like 2 isoform 1 n=1 T... 55 2e-06
UniRef100_UPI0000E1F765 PREDICTED: formin-like 2 isoform 7 n=1 T... 55 2e-06
UniRef100_UPI0000E1F763 PREDICTED: formin-like 2 isoform 4 n=1 T... 55 2e-06
UniRef100_UPI0000D5650F PREDICTED: similar to GCR(ich) CG5812-PA... 55 2e-06
UniRef100_UPI0000025476 DEAH (Asp-Glu-Ala-His) box polypeptide 9... 55 2e-06
UniRef100_UPI00001EDBAC DEAH (Asp-Glu-Ala-His) box polypeptide 9... 55 2e-06
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 55 2e-06
UniRef100_B6ID90 DEAH (Asp-Glu-Ala-His) box polypeptide 9 (Fragm... 55 2e-06
UniRef100_Q89FZ1 Bll6557 protein n=1 Tax=Bradyrhizobium japonicu... 55 2e-06
UniRef100_Q46I49 RNA-binding region RNP-1 (RNA recognition motif... 55 2e-06
UniRef100_Q2W222 RTX toxins and related Ca2+-binding protein n=1... 55 2e-06
UniRef100_D0CMI9 RNA-binding region RNP-1 n=1 Tax=Synechococcus ... 55 2e-06
UniRef100_B7QTD0 Single-stranded DNA-binding protein n=1 Tax=Rue... 55 2e-06
UniRef100_Q9SIH2 Putative uncharacterized protein At2g36120 n=1 ... 55 2e-06
UniRef100_Q9LW52 Genomic DNA, chromosome 3, P1 clone: MLM24 n=1 ... 55 2e-06
UniRef100_Q6H3Y0 Os02g0651900 protein n=1 Tax=Oryza sativa Japon... 55 2e-06
UniRef100_Q41187 Glycine-rich protein (Fragment) n=1 Tax=Arabido... 55 2e-06
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 55 2e-06
UniRef100_C7IZY4 Os03g0189900 protein n=1 Tax=Oryza sativa Japon... 55 2e-06
UniRef100_C5XW81 Putative uncharacterized protein Sb04g005115 n=... 55 2e-06
UniRef100_B2BXJ4 LTRGag-pol-polymerase 3 n=1 Tax=Arabidopsis lyr... 55 2e-06
UniRef100_A3BK86 Putative uncharacterized protein n=1 Tax=Oryza ... 55 2e-06
UniRef100_Q9VB86 TweedleT n=1 Tax=Drosophila melanogaster RepID=... 55 2e-06
UniRef100_Q9U1G9 Gly-rich protein n=1 Tax=Drosophila melanogaste... 55 2e-06
UniRef100_Q0IGW9 LP21747p n=1 Tax=Drosophila melanogaster RepID=... 55 2e-06
UniRef100_B7PS04 Putative uncharacterized protein (Fragment) n=1... 55 2e-06
UniRef100_B5DM33 GA28222 n=1 Tax=Drosophila pseudoobscura pseudo... 55 2e-06
UniRef100_B5DJN5 GA28844 n=1 Tax=Drosophila pseudoobscura pseudo... 55 2e-06
UniRef100_B4P4K6 GE13380 n=1 Tax=Drosophila yakuba RepID=B4P4K6_... 55 2e-06
UniRef100_B4NC69 GK25802 n=1 Tax=Drosophila willistoni RepID=B4N... 55 2e-06
UniRef100_B4M6Y6 GJ16553 n=1 Tax=Drosophila virilis RepID=B4M6Y6... 55 2e-06
UniRef100_B4M1Y2 GJ18769 n=1 Tax=Drosophila virilis RepID=B4M1Y2... 55 2e-06
UniRef100_B4JNX2 GH24906 n=1 Tax=Drosophila grimshawi RepID=B4JN... 55 2e-06
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 55 2e-06
UniRef100_B4IEZ2 GM13499 n=1 Tax=Drosophila sechellia RepID=B4IE... 55 2e-06
UniRef100_B3N5L2 GG24240 n=1 Tax=Drosophila erecta RepID=B3N5L2_... 55 2e-06
UniRef100_A9V3V4 Predicted protein n=1 Tax=Monosiga brevicollis ... 55 2e-06
UniRef100_Q9P6T1 Putative uncharacterized protein 15E6.220 n=1 T... 55 2e-06
UniRef100_B0CVI1 Metacaspase n=1 Tax=Laccaria bicolor S238N-H82 ... 55 2e-06
UniRef100_A7UWD4 Predicted protein n=1 Tax=Neurospora crassa Rep... 55 2e-06
UniRef100_P12978 Epstein-Barr nuclear antigen 2 n=1 Tax=Human he... 55 2e-06
UniRef100_A1DGZ7 ATP-dependent RNA helicase dbp2 n=1 Tax=Neosart... 55 2e-06
UniRef100_UPI0001837EA1 hypothetical protein LOC100130302 n=1 Ta... 55 3e-06
UniRef100_B7XDE6 Glycoprotein n=1 Tax=Equid herpesvirus 9 RepID=... 55 3e-06
UniRef100_B7FED2 Envelope glycoprotein n=1 Tax=Equid herpesvirus... 55 3e-06
UniRef100_B9N9N3 Predicted protein n=1 Tax=Populus trichocarpa R... 55 3e-06
UniRef100_C5LQI8 Facilitator of iron transport 3, putative n=1 T... 55 3e-06
UniRef100_B4N2N2 GK24882 n=1 Tax=Drosophila willistoni RepID=B4N... 55 3e-06
UniRef100_A8NK60 Antifreeze peptide 4, putative n=1 Tax=Brugia m... 55 3e-06
UniRef100_UPI0001A7B366 GRP23 (GLYCINE-RICH PROTEIN 23) n=1 Tax=... 55 3e-06
UniRef100_UPI0000DB7618 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0000DA1EB9 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI00006A18B5 UPI00006A18B5 related cluster n=1 Tax=Xe... 55 3e-06
UniRef100_UPI00006A12E4 Keratin 13. n=1 Tax=Xenopus (Silurana) t... 55 3e-06
UniRef100_UPI000069FBE4 Formin-like protein 2 (Formin homology 2... 55 3e-06
UniRef100_UPI000069FBE3 Formin-like protein 2 (Formin homology 2... 55 3e-06
UniRef100_UPI000179CB3D inverted formin 2 isoform 1 n=1 Tax=Bos ... 55 3e-06
UniRef100_Q5M8X9 Keratin 13 n=1 Tax=Xenopus (Silurana) tropicali... 55 3e-06
UniRef100_Q65553 UL36 n=3 Tax=Bovine herpesvirus 1 RepID=Q65553_... 55 3e-06
UniRef100_B9EK12 Keratin associated protein 16-7 n=1 Tax=Mus mus... 55 3e-06
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 55 3e-06
UniRef100_Q9MBF4 Glycine-rich protein n=1 Tax=Citrus unshiu RepI... 55 3e-06
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 55 3e-06
UniRef100_Q2QV63 Os12g0242100 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_Q10FE7 Os03g0670700 protein n=1 Tax=Oryza sativa Japon... 55 3e-06
UniRef100_Q0WQG4 Putative uncharacterized protein At1g05135 (Fra... 55 3e-06
UniRef100_C5YNX7 Putative uncharacterized protein Sb08g015580 n=... 55 3e-06
UniRef100_B9SS24 Glycine-rich cell wall structural protein 1.8, ... 55 3e-06
UniRef100_A4S3M6 Predicted protein n=1 Tax=Ostreococcus lucimari... 55 3e-06
UniRef100_A3BDN5 Putative uncharacterized protein n=1 Tax=Oryza ... 55 3e-06
UniRef100_Q9XUW5 Protein F58E10.3a, confirmed by transcript evid... 55 3e-06
UniRef100_B4JLJ8 GH12877 n=1 Tax=Drosophila grimshawi RepID=B4JL... 55 3e-06
UniRef100_A7URV0 AGAP006973-PA n=1 Tax=Anopheles gambiae RepID=A... 55 3e-06
UniRef100_A4HT33 ATP-dependent DEAD/H RNA helicase, putative n=1... 55 3e-06
UniRef100_B2W8S4 Metacaspase n=1 Tax=Pyrenophora tritici-repenti... 55 3e-06
UniRef100_P17130-2 Isoform A1-B of Heterogeneous nuclear ribonuc... 55 3e-06
UniRef100_P17130 Heterogeneous nuclear ribonucleoproteins A1 hom... 55 3e-06
UniRef100_Q925H4 Keratin-associated protein 16-7 n=1 Tax=Mus mus... 55 3e-06
UniRef100_P10495 Glycine-rich cell wall structural protein 1.0 n... 55 3e-06
UniRef100_P19470 Eggshell protein 1 n=1 Tax=Schistosoma japonicu... 55 3e-06
UniRef100_A4QSS5 ATP-dependent RNA helicase DBP2 n=1 Tax=Magnapo... 55 3e-06
UniRef100_P08826 Chorion class A protein L11 n=1 Tax=Bombyx mori... 55 3e-06
UniRef100_Q825Z2 Putative proline-rich protein n=1 Tax=Streptomy... 49 3e-06
UniRef100_A8R200 ALEX (Fragment) n=1 Tax=Apodemus agrarius RepID... 54 4e-06
UniRef100_A4YN54 Putative uncharacterized protein n=1 Tax=Bradyr... 54 4e-06
UniRef100_A0LTP3 Transcriptional regulators, TraR/DksA family n=... 54 4e-06
UniRef100_C9KI85 Ribonuclease, Rne/Rng family n=1 Tax=Sanguibact... 54 4e-06
UniRef100_C1E5Q6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 4e-06
UniRef100_Q4QEM8 Putative uncharacterized protein n=1 Tax=Leishm... 54 4e-06
UniRef100_UPI0001982C16 PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI00017916EE PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI00016C5334 RNA-binding region RNP-1 n=1 Tax=Gemmata... 54 4e-06
UniRef100_UPI0000E80902 PREDICTED: similar to RAPH1 protein n=1 ... 54 4e-06
UniRef100_UPI0000DA3CD5 PREDICTED: hypothetical protein n=1 Tax=... 54 4e-06
UniRef100_UPI000069F4F8 Heterogeneous nuclear ribonucleoprotein ... 54 4e-06
UniRef100_Q90WR5 Keratin alpha n=1 Tax=Lampetra fluviatilis RepI... 54 4e-06
UniRef100_B2GUG7 Krt5.4 protein n=1 Tax=Xenopus (Silurana) tropi... 54 4e-06
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 54 4e-06
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 54 4e-06
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 54 4e-06
UniRef100_Q8RUS0 Putative uncharacterized protein At2g18115 n=1 ... 54 4e-06
UniRef100_Q8GTL0 Putative glycine-rich cell wall protein n=2 Tax... 54 4e-06
UniRef100_Q53N23 Transposon protein, putative, CACTA, En/Spm sub... 54 4e-06
UniRef100_Q40052 Glycine rich protein, RNA binding protein n=1 T... 54 4e-06
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 54 4e-06
UniRef100_C5XP48 Putative uncharacterized protein Sb03g005056 (F... 54 4e-06
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 54 4e-06
UniRef100_B9T304 Glycine-rich cell wall structural protein 1, pu... 54 4e-06
UniRef100_B9SVA0 GRP3S, putative n=1 Tax=Ricinus communis RepID=... 54 4e-06
UniRef100_B9GME0 Predicted protein n=1 Tax=Populus trichocarpa R... 54 4e-06
UniRef100_B8BP19 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_B6TZT1 Glycine-rich cell wall structural protein n=1 T... 54 4e-06
UniRef100_A9NNT8 Putative uncharacterized protein n=1 Tax=Picea ... 54 4e-06
UniRef100_A9NML8 Putative uncharacterized protein n=1 Tax=Picea ... 54 4e-06
UniRef100_A2YQ51 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_A2XBH9 Putative uncharacterized protein n=1 Tax=Oryza ... 54 4e-06
UniRef100_Q868B4 Protein ZK643.8, partially confirmed by transcr... 54 4e-06
UniRef100_Q7JRD4 CG4663 n=1 Tax=Drosophila melanogaster RepID=Q7... 54 4e-06
UniRef100_C4WUK3 ACYPI005604 protein n=1 Tax=Acyrthosiphon pisum... 54 4e-06