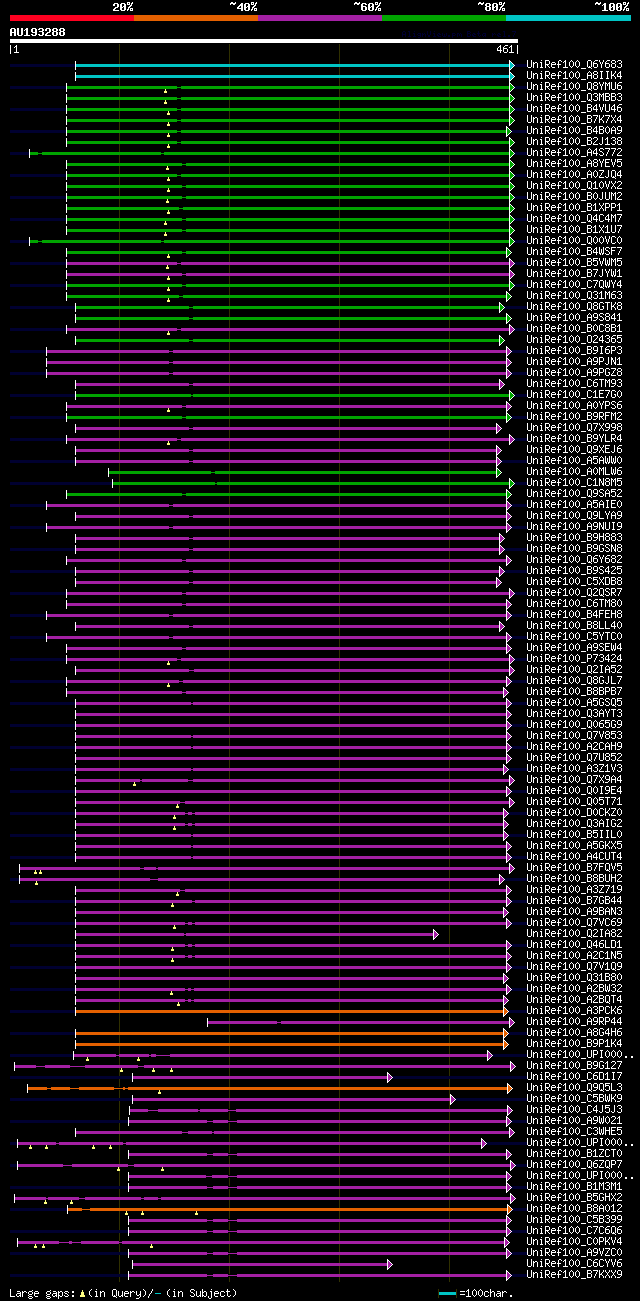
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AU193288 PFL043b12_r
(461 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q6Y683 41 kDa ribosome-associated protein n=1 Tax=Chla... 270 4e-71
UniRef100_A8IIK4 Chloroplast stem-loop-binding protein n=1 Tax=C... 270 4e-71
UniRef100_Q8YMU6 mRNA-binding protein n=1 Tax=Nostoc sp. PCC 712... 132 9e-30
UniRef100_Q3MBB3 3-beta hydroxysteroid dehydrogenase/isomerase n... 132 9e-30
UniRef100_B4VU46 3-beta hydroxysteroid dehydrogenase/isomerase f... 130 3e-29
UniRef100_B7K7X4 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 130 6e-29
UniRef100_B4B0A9 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 130 6e-29
UniRef100_B2J138 NAD-dependent epimerase/dehydratase n=1 Tax=Nos... 128 2e-28
UniRef100_A4S772 Predicted protein (Fragment) n=1 Tax=Ostreococc... 128 2e-28
UniRef100_A8YEV5 Genome sequencing data, contig C301 n=1 Tax=Mic... 128 2e-28
UniRef100_A0ZJQ4 3-beta hydroxysteroid dehydrogenase/isomerase n... 127 3e-28
UniRef100_Q10VX2 NAD-dependent epimerase/dehydratase n=1 Tax=Tri... 126 7e-28
UniRef100_B0JUM2 NAD-dependent epimerase/dehydratase n=1 Tax=Mic... 126 9e-28
UniRef100_B1XPP1 NAD dependent epimerase/dehydratase family prot... 125 1e-27
UniRef100_Q4C4M7 Similar to Nucleoside-diphosphate-sugar epimera... 124 3e-27
UniRef100_B1X1U7 mRNA-binding protein n=1 Tax=Cyanothece sp. ATC... 122 1e-26
UniRef100_Q00VC0 PREDICTED OJ1664_D08.105 gene product (ISS) n=1... 122 1e-26
UniRef100_B4WSF7 3-beta hydroxysteroid dehydrogenase/isomerase f... 122 2e-26
UniRef100_B5VWM5 NAD-dependent epimerase/dehydratase n=1 Tax=Art... 121 3e-26
UniRef100_B7JYW1 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 119 1e-25
UniRef100_C7QWY4 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 119 1e-25
UniRef100_Q31M63 mRNA-binding protein n=2 Tax=Synechococcus elon... 119 1e-25
UniRef100_Q8GTK8 Os07g0212200 protein n=2 Tax=Oryza sativa RepID... 119 1e-25
UniRef100_A9S841 Predicted protein n=1 Tax=Physcomitrella patens... 119 1e-25
UniRef100_B0C8B1 NAD-dependent epimerase/dehydratase family prot... 118 2e-25
UniRef100_O24365 Chloroplast mRNA-binding protein CSP41 (Fragmen... 118 2e-25
UniRef100_B9I6P3 Predicted protein n=1 Tax=Populus trichocarpa R... 117 4e-25
UniRef100_A9PJN1 Putative uncharacterized protein n=1 Tax=Populu... 117 4e-25
UniRef100_A9PGZ8 Putative uncharacterized protein n=1 Tax=Populu... 117 4e-25
UniRef100_C6TM93 Putative uncharacterized protein n=1 Tax=Glycin... 116 7e-25
UniRef100_C1E7G0 Predicted protein n=1 Tax=Micromonas sp. RCC299... 115 1e-24
UniRef100_A0YPS6 3-beta hydroxysteroid dehydrogenase/isomerase n... 115 2e-24
UniRef100_B9RFM2 NAD dependent epimerase/dehydratase, putative n... 115 2e-24
UniRef100_Q7X998 MRNA-binding protein (Fragment) n=1 Tax=Nicotia... 114 3e-24
UniRef100_B9YLR4 NAD-dependent epimerase/dehydratase n=1 Tax='No... 114 4e-24
UniRef100_Q9XEJ6 MRNA binding protein n=1 Tax=Solanum lycopersic... 114 4e-24
UniRef100_A5AWW0 Chromosome chr7 scaffold_31, whole genome shotg... 114 4e-24
UniRef100_A0MLW6 MRNA-binding protein (Fragment) n=1 Tax=Capsicu... 114 4e-24
UniRef100_C1N8M5 Predicted protein n=1 Tax=Micromonas pusilla CC... 113 6e-24
UniRef100_Q9SA52 Uncharacterized protein At1g09340, chloroplasti... 113 6e-24
UniRef100_A5AIE0 Chromosome chr14 scaffold_27, whole genome shot... 112 1e-23
UniRef100_Q9LYA9 Uncharacterized protein At3g63140, chloroplasti... 112 1e-23
UniRef100_A9NUI9 Putative uncharacterized protein n=1 Tax=Picea ... 112 2e-23
UniRef100_B9H883 Predicted protein (Fragment) n=1 Tax=Populus tr... 111 2e-23
UniRef100_B9GSN8 Predicted protein n=1 Tax=Populus trichocarpa R... 111 2e-23
UniRef100_Q6Y682 38 kDa ribosome-associated protein n=1 Tax=Chla... 110 4e-23
UniRef100_B9S425 NAD dependent epimerase/dehydratase, putative n... 110 4e-23
UniRef100_C5XDB8 Putative uncharacterized protein Sb02g006430 n=... 110 5e-23
UniRef100_Q2QSR7 Os12g0420200 protein n=1 Tax=Oryza sativa Japon... 109 8e-23
UniRef100_C6TM80 Putative uncharacterized protein n=1 Tax=Glycin... 108 2e-22
UniRef100_B4FEH8 Putative uncharacterized protein n=1 Tax=Zea ma... 108 2e-22
UniRef100_B8LL40 Putative uncharacterized protein n=1 Tax=Picea ... 107 3e-22
UniRef100_C5YTC0 Putative uncharacterized protein Sb08g005500 n=... 107 5e-22
UniRef100_A9SEW4 Predicted protein n=1 Tax=Physcomitrella patens... 105 2e-21
UniRef100_P73424 Slr1540 protein n=1 Tax=Synechocystis sp. PCC 6... 104 3e-21
UniRef100_Q2IA52 Chloroplast mRNA binding protein csp41 n=1 Tax=... 102 1e-20
UniRef100_Q8GJL7 Putative uncharacterized protein SEM0023 n=1 Ta... 98 3e-19
UniRef100_B8BPB7 Putative uncharacterized protein n=1 Tax=Oryza ... 96 1e-18
UniRef100_A5GSQ5 NAD dependent epimerase/dehydratase n=1 Tax=Syn... 94 4e-18
UniRef100_Q3AYT3 Possible nucleotide sugar epimerase n=1 Tax=Syn... 93 1e-17
UniRef100_Q065G9 Possible nucleotide sugar epimerase n=1 Tax=Syn... 92 2e-17
UniRef100_Q7V853 Possible mRNA-binding protein n=1 Tax=Prochloro... 91 3e-17
UniRef100_A2CAH9 Possible mRNA binding protein n=1 Tax=Prochloro... 91 3e-17
UniRef100_Q7U852 Possible nucleotide sugar epimerase n=1 Tax=Syn... 91 5e-17
UniRef100_A3Z1V3 Possible nucleotide sugar epimerase n=1 Tax=Syn... 90 9e-17
UniRef100_Q7X9A4 MRNA binding protein (Fragment) n=1 Tax=Bigelow... 89 1e-16
UniRef100_Q0I9E4 Possible nucleotide sugar epimerase n=1 Tax=Syn... 89 2e-16
UniRef100_Q05T71 Possible nucleotide sugar epimerase n=1 Tax=Syn... 89 2e-16
UniRef100_D0CKZ0 Possible nucleotide sugar epimerase n=1 Tax=Syn... 89 2e-16
UniRef100_Q3AIG2 Possible nucleotide sugar epimerase n=1 Tax=Syn... 88 3e-16
UniRef100_B5IIL0 Possible nucleotide sugar epimerase n=1 Tax=Cya... 87 4e-16
UniRef100_A5GKX5 NAD dependent epimerase/dehydratase n=1 Tax=Syn... 87 8e-16
UniRef100_A4CUT4 Possible nucleotide sugar epimerase n=1 Tax=Syn... 85 3e-15
UniRef100_B7FQV5 Predicted protein n=1 Tax=Phaeodactylum tricorn... 84 5e-15
UniRef100_B8BUH2 Predicted protein n=1 Tax=Thalassiosira pseudon... 81 3e-14
UniRef100_A3Z719 Possible mRNA-binding protein n=1 Tax=Synechoco... 79 2e-13
UniRef100_B7GB44 Predicted protein n=1 Tax=Phaeodactylum tricorn... 79 2e-13
UniRef100_A9BAN3 Possible mRNA binding protein n=1 Tax=Prochloro... 78 3e-13
UniRef100_Q7VC69 NAD dependent epimerase/dehydratase n=1 Tax=Pro... 77 6e-13
UniRef100_Q2IA82 Chloroplast mRNA binding protein csp41 (Fragmen... 76 1e-12
UniRef100_Q46LD1 Possible mRNA-binding protein n=1 Tax=Prochloro... 75 3e-12
UniRef100_A2C1N5 Possible mRNA binding protein n=1 Tax=Prochloro... 74 5e-12
UniRef100_Q7V1Q9 Possible mRNA binding protein n=1 Tax=Prochloro... 64 4e-09
UniRef100_Q31B80 mRNA binding protein-like protein n=1 Tax=Proch... 62 3e-08
UniRef100_A2BW32 Possible mRNA binding protein n=1 Tax=Prochloro... 61 3e-08
UniRef100_A2BQT4 Possible mRNA binding protein n=1 Tax=Prochloro... 61 4e-08
UniRef100_A3PCK6 Possible mRNA binding protein n=1 Tax=Prochloro... 60 1e-07
UniRef100_A9RP44 Predicted protein n=1 Tax=Physcomitrella patens... 60 1e-07
UniRef100_A8G4H6 Possible mRNA binding protein n=1 Tax=Prochloro... 59 1e-07
UniRef100_B9P1K4 NAD dependent epimerase/dehydratase n=1 Tax=Pro... 59 1e-07
UniRef100_UPI0000E7FCDE PREDICTED: frizzled homolog 8 (Drosophil... 59 2e-07
UniRef100_B9G127 Putative uncharacterized protein n=1 Tax=Oryza ... 59 2e-07
UniRef100_C6D1I7 NAD-dependent epimerase/dehydratase n=1 Tax=Pae... 58 3e-07
UniRef100_Q9Q5L3 EBNA-2 n=1 Tax=Macacine herpesvirus 4 RepID=Q9Q... 57 5e-07
UniRef100_C5BWK9 NAD-dependent epimerase/dehydratase n=1 Tax=Beu... 57 5e-07
UniRef100_C4J5J3 Putative uncharacterized protein n=1 Tax=Zea ma... 56 1e-06
UniRef100_A9W021 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 56 1e-06
UniRef100_C3WHE5 Isoflavone reductase n=1 Tax=Fusobacterium sp. ... 56 1e-06
UniRef100_UPI0000DD95B9 Os08g0528700 n=1 Tax=Oryza sativa Japoni... 55 2e-06
UniRef100_B1ZCT0 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 55 2e-06
UniRef100_Q6ZQP7 Uncharacterized protein LOC284861 n=1 Tax=Homo ... 55 2e-06
UniRef100_UPI000038279A COG0451: Nucleoside-diphosphate-sugar ep... 55 2e-06
UniRef100_B1M3M1 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 55 2e-06
UniRef100_B5GHX2 Predicted protein n=1 Tax=Streptomyces sp. SPB7... 55 2e-06
UniRef100_B8A012 Putative uncharacterized protein n=1 Tax=Zea ma... 55 2e-06
UniRef100_C5B399 Putative UDP-glucose 4-epimerase n=1 Tax=Methyl... 55 3e-06
UniRef100_C7C6Q6 Putative UDP-glucose 4-epimerase n=1 Tax=Methyl... 55 3e-06
UniRef100_C0PKV4 Putative uncharacterized protein n=1 Tax=Zea ma... 55 3e-06
UniRef100_A9VZC0 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 54 4e-06
UniRef100_C6CYV6 NAD-dependent epimerase/dehydratase n=1 Tax=Pae... 53 9e-06
UniRef100_B7KXX9 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 53 9e-06
UniRef100_A9GTF5 NDP-sugar oxidoreductase similar to UDP-glucose... 53 9e-06