[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
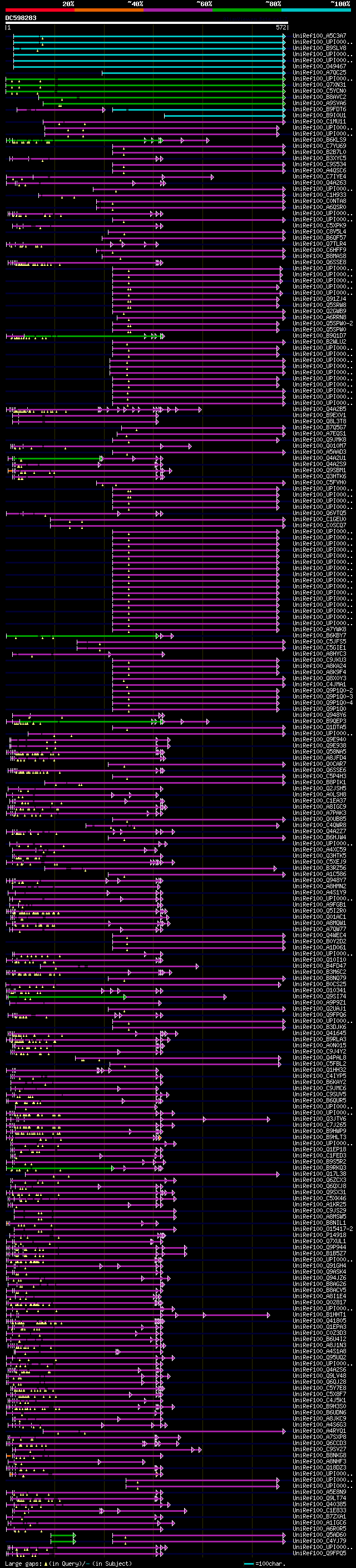
Query= DC598283 MPDL063b04_r
(572 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_A5C3A7 Putative uncharacterized protein n=1 Tax=Vitis ... 258 2e-67
UniRef100_UPI0001984296 PREDICTED: hypothetical protein n=1 Tax=... 257 5e-67
UniRef100_B9SLV8 Vacuolar protein sorting, putative n=1 Tax=Rici... 246 9e-64
UniRef100_UPI00017393D2 protein binding n=1 Tax=Arabidopsis thal... 234 3e-60
UniRef100_UPI0000196D1B protein binding n=1 Tax=Arabidopsis thal... 234 3e-60
UniRef100_O49467 Putative uncharacterized protein AT4g19490 n=1 ... 234 3e-60
UniRef100_A7QC25 Chromosome chr10 scaffold_76, whole genome shot... 212 1e-53
UniRef100_UPI0000DD8F7C Os04g0212100 n=1 Tax=Oryza sativa Japoni... 191 3e-47
UniRef100_Q7XN31 OSJNBa0083I11.13 protein n=1 Tax=Oryza sativa J... 191 3e-47
UniRef100_C5YCN0 Putative uncharacterized protein Sb06g002040 n=... 191 4e-47
UniRef100_B8AVC2 Putative uncharacterized protein n=1 Tax=Oryza ... 191 4e-47
UniRef100_A9SVA6 Predicted protein n=1 Tax=Physcomitrella patens... 174 3e-42
UniRef100_B9FDT6 Putative uncharacterized protein n=1 Tax=Oryza ... 144 4e-35
UniRef100_B9I0U1 Predicted protein n=1 Tax=Populus trichocarpa R... 130 9e-29
UniRef100_C1MU11 Predicted protein n=1 Tax=Micromonas pusilla CC... 87 7e-16
UniRef100_UPI0000E47B88 PREDICTED: similar to tumor antigen SLP-... 87 9e-16
UniRef100_UPI0000E46A02 PREDICTED: similar to vacuolar sorting p... 87 9e-16
UniRef100_B6KLS9 Voltage gated chloride channel domain-containin... 83 2e-14
UniRef100_C7YU69 Predicted protein n=1 Tax=Nectria haematococca ... 83 2e-14
UniRef100_B2B7L0 Predicted CDS Pa_2_11470 n=1 Tax=Podospora anse... 83 2e-14
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 82 2e-14
UniRef100_C9S534 Vacuolar protein sorting-associated protein n=1... 82 2e-14
UniRef100_A4QSC6 Putative uncharacterized protein n=1 Tax=Magnap... 82 4e-14
UniRef100_C7IYE4 Os02g0557000 protein (Fragment) n=1 Tax=Oryza s... 81 5e-14
UniRef100_Q4A263 Putative membrane protein n=1 Tax=Emiliania hux... 81 6e-14
UniRef100_UPI0001861CD3 hypothetical protein BRAFLDRAFT_213066 n... 80 8e-14
UniRef100_C1H933 Putative uncharacterized protein n=1 Tax=Paraco... 80 8e-14
UniRef100_C0NTA8 Vacuolar sorting-associated protein n=1 Tax=Aje... 80 8e-14
UniRef100_A6QSR0 Putative uncharacterized protein n=1 Tax=Ajello... 80 8e-14
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 80 1e-13
UniRef100_UPI000023F2D4 hypothetical protein FG06778.1 n=1 Tax=G... 80 1e-13
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 80 1e-13
UniRef100_C8V5L4 GARP complex component (Vps54), putative (AFU_o... 80 1e-13
UniRef100_B6QF57 GARP complex component (Vps54), putative n=1 Ta... 80 1e-13
UniRef100_Q7TLR4 Putative uncharacterized protein n=1 Tax=Choris... 80 1e-13
UniRef100_C6HFF9 Vacuolar sorting protein n=1 Tax=Ajellomyces ca... 79 2e-13
UniRef100_B8MAS8 GARP complex component (Vps54), putative n=1 Ta... 79 2e-13
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 79 2e-13
UniRef100_UPI000194C002 PREDICTED: vacuolar protein sorting 54 h... 79 3e-13
UniRef100_UPI0000E7FFFE PREDICTED: similar to vacuolar sorting p... 79 3e-13
UniRef100_UPI0000E7FFFD PREDICTED: similar to vacuolar sorting p... 79 3e-13
UniRef100_UPI00015DEBD5 vacuolar protein sorting 54 (yeast) n=1 ... 79 3e-13
UniRef100_UPI0000ECC93A Vacuolar protein sorting-associated prot... 79 3e-13
UniRef100_Q91ZJ4 Hcc8 n=1 Tax=Mus musculus RepID=Q91ZJ4_MOUSE 79 3e-13
UniRef100_Q5SRW8 Vacuolar protein sorting 54 (Yeast) (Fragment) ... 79 3e-13
UniRef100_Q2GWB9 Putative uncharacterized protein n=1 Tax=Chaeto... 79 3e-13
UniRef100_A6RRN8 Putative uncharacterized protein n=1 Tax=Botryo... 79 3e-13
UniRef100_Q5SPW0-2 Isoform 2 of Vacuolar protein sorting-associa... 79 3e-13
UniRef100_Q5SPW0 Vacuolar protein sorting-associated protein 54 ... 79 3e-13
UniRef100_B9Q1D7 Chloride channel, putative n=1 Tax=Toxoplasma g... 78 4e-13
UniRef100_B2WLU2 Putative uncharacterized protein n=1 Tax=Pyreno... 78 4e-13
UniRef100_UPI000155D0B7 PREDICTED: similar to vacuolar sorting p... 78 5e-13
UniRef100_UPI000155D0B6 PREDICTED: similar to vacuolar sorting p... 78 5e-13
UniRef100_UPI00006A0C1B Vacuolar protein sorting-associated prot... 78 5e-13
UniRef100_UPI00006A0C1A Vacuolar protein sorting-associated prot... 78 5e-13
UniRef100_UPI00006A0C19 Vacuolar protein sorting-associated prot... 78 5e-13
UniRef100_UPI0001B7A932 Vacuolar protein sorting-associated prot... 78 5e-13
UniRef100_UPI000019BA10 Vacuolar protein sorting-associated prot... 78 5e-13
UniRef100_UPI00016E4FF6 UPI00016E4FF6 related cluster n=1 Tax=Ta... 78 5e-13
UniRef100_UPI00016E4DD0 UPI00016E4DD0 related cluster n=1 Tax=Ta... 78 5e-13
UniRef100_UPI00016E4DCF UPI00016E4DCF related cluster n=1 Tax=Ta... 78 5e-13
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 78 5e-13
UniRef100_B9EXV1 Putative uncharacterized protein n=1 Tax=Oryza ... 78 5e-13
UniRef100_Q8L3T8 cDNA clone:001-201-A04, full insert sequence n=... 78 5e-13
UniRef100_B7Q5G7 Vacuolar protein-sorting protein, putative n=1 ... 78 5e-13
UniRef100_A7EQS1 Putative uncharacterized protein n=1 Tax=Sclero... 78 5e-13
UniRef100_Q9JMK8 Vacuolar protein sorting-associated protein 54 ... 78 5e-13
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 77 7e-13
UniRef100_A5AAD3 Complex: S. cerevisiae Vps52p n=1 Tax=Aspergill... 77 7e-13
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 77 9e-13
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 77 1e-12
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 77 1e-12
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 77 1e-12
UniRef100_C5FVH0 Putative uncharacterized protein n=1 Tax=Micros... 77 1e-12
UniRef100_UPI000155F786 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI000155F785 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI0000F2B5CB PREDICTED: similar to vacuolar sorting p... 76 2e-12
UniRef100_UPI0000F2B5CA PREDICTED: similar to vacuolar sorting p... 76 2e-12
UniRef100_Q6VTQ5 Putative uncharacterized protein n=1 Tax=Choris... 76 2e-12
UniRef100_C1GEU0 Putative uncharacterized protein n=1 Tax=Paraco... 76 2e-12
UniRef100_C0SCQ7 Putative uncharacterized protein n=1 Tax=Paraco... 76 2e-12
UniRef100_UPI0000E1F406 PREDICTED: vacuolar protein sorting 54 i... 76 2e-12
UniRef100_UPI0000E1F405 PREDICTED: vacuolar protein sorting 54 i... 76 2e-12
UniRef100_UPI0000E1F404 PREDICTED: vacuolar protein sorting 54 i... 76 2e-12
UniRef100_UPI0000E1F403 PREDICTED: vacuolar protein sorting 54 i... 76 2e-12
UniRef100_UPI0000D9D458 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI0000D9D457 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI0000D9D456 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI0000D9D455 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI00005A21FA PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI00005A21F8 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI00004BB762 PREDICTED: similar to vacuolar protein s... 76 2e-12
UniRef100_UPI0000EB2BE7 Vacuolar protein sorting-associated prot... 76 2e-12
UniRef100_UPI00004BB761 Vacuolar protein sorting-associated prot... 76 2e-12
UniRef100_UPI000179E68A UPI000179E68A related cluster n=1 Tax=Bo... 76 2e-12
UniRef100_UPI000179E67D UPI000179E67D related cluster n=1 Tax=Bo... 76 2e-12
UniRef100_UPI0000F32C72 UPI0000F32C72 related cluster n=1 Tax=Bo... 76 2e-12
UniRef100_A7YWK8 VPS54 protein n=1 Tax=Bos taurus RepID=A7YWK8_B... 76 2e-12
UniRef100_B6KBY7 Putative uncharacterized protein n=1 Tax=Toxopl... 76 2e-12
UniRef100_C5JFS5 GARP complex component n=1 Tax=Ajellomyces derm... 76 2e-12
UniRef100_C5GIE1 GARP complex component n=1 Tax=Ajellomyces derm... 76 2e-12
UniRef100_A8HYC3 Hydroxyproline-rich glycoprotein n=1 Tax=Chlamy... 75 3e-12
UniRef100_C9JKU3 Putative uncharacterized protein VPS54 n=1 Tax=... 75 3e-12
UniRef100_A8KA24 cDNA FLJ78648, highly similar to Homo sapiens v... 75 3e-12
UniRef100_A8K9F4 cDNA FLJ77625, highly similar to Homo sapiens v... 75 3e-12
UniRef100_Q8X0Y3 Putative uncharacterized protein n=1 Tax=Neuros... 75 3e-12
UniRef100_C4JMA1 Putative uncharacterized protein n=1 Tax=Uncino... 75 3e-12
UniRef100_Q9P1Q0-2 Isoform 2 of Vacuolar protein sorting-associa... 75 3e-12
UniRef100_Q9P1Q0-3 Isoform 3 of Vacuolar protein sorting-associa... 75 3e-12
UniRef100_Q9P1Q0-4 Isoform 4 of Vacuolar protein sorting-associa... 75 3e-12
UniRef100_Q9P1Q0 Vacuolar protein sorting-associated protein 54 ... 75 3e-12
UniRef100_Q948Y6 VMP4 protein n=1 Tax=Volvox carteri f. nagarien... 75 5e-12
UniRef100_B9QEP3 Chloride channel protein k, putative n=1 Tax=To... 75 5e-12
UniRef100_Q1DTA5 Putative uncharacterized protein n=1 Tax=Coccid... 75 5e-12
UniRef100_UPI00017B4E2A UPI00017B4E2A related cluster n=1 Tax=Te... 74 6e-12
UniRef100_Q9E940 ICP4 protein n=1 Tax=Gallid herpesvirus 3 RepID... 74 6e-12
UniRef100_Q9E938 ICP4 protein n=1 Tax=Gallid herpesvirus 3 RepID... 74 6e-12
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 74 6e-12
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 74 6e-12
UniRef100_Q0CAR7 Putative uncharacterized protein n=1 Tax=Asperg... 74 6e-12
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 74 8e-12
UniRef100_C5P4H3 Putative uncharacterized protein n=1 Tax=Coccid... 74 8e-12
UniRef100_B8PIK1 Predicted protein n=1 Tax=Postia placenta Mad-6... 74 8e-12
UniRef100_Q2JSH5 Protein kinase n=1 Tax=Synechococcus sp. JA-3-3... 74 1e-11
UniRef100_A0LSH8 Glycoside hydrolase, family 6 n=1 Tax=Acidother... 74 1e-11
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 74 1e-11
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 74 1e-11
UniRef100_A7PAK3 Chromosome chr14 scaffold_9, whole genome shotg... 74 1e-11
UniRef100_Q0UB85 Putative uncharacterized protein n=1 Tax=Phaeos... 74 1e-11
UniRef100_C4QWR8 Component of the GARP (Golgi-associated retrogr... 74 1e-11
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 73 1e-11
UniRef100_B6HJW4 Pc21g15660 protein n=1 Tax=Penicillium chrysoge... 73 1e-11
UniRef100_UPI0000E12B23 Os07g0496300 n=1 Tax=Oryza sativa Japoni... 73 2e-11
UniRef100_A4XC59 NLP/P60 protein n=1 Tax=Salinispora tropica CNB... 73 2e-11
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 73 2e-11
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 73 2e-11
UniRef100_B3RZ56 Putative uncharacterized protein (Fragment) n=1... 73 2e-11
UniRef100_A1C586 GARP complex component (Vps54), putative n=1 Ta... 73 2e-11
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 72 2e-11
UniRef100_A8HMN2 Predicted protein n=1 Tax=Chlamydomonas reinhar... 72 2e-11
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 72 2e-11
UniRef100_UPI0001982E0B PREDICTED: hypothetical protein n=1 Tax=... 72 3e-11
UniRef100_A9FGB1 Putative uncharacterized protein n=1 Tax=Sorang... 72 3e-11
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 72 3e-11
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 72 3e-11
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 72 3e-11
UniRef100_A7QW77 Chromosome chr3 scaffold_199, whole genome shot... 72 3e-11
UniRef100_Q4WEC4 GARP complex component (Vps54), putative n=1 Ta... 72 3e-11
UniRef100_B0Y2D2 GARP complex component (Vps54), putative n=1 Ta... 72 3e-11
UniRef100_A1D061 GARP complex component (Vps54), putative n=1 Ta... 72 3e-11
UniRef100_UPI0000DA2535 PREDICTED: hypothetical protein n=1 Tax=... 72 4e-11
UniRef100_Q10I10 Os03g0568800 protein n=1 Tax=Oryza sativa Japon... 72 4e-11
UniRef100_B4FD47 Putative uncharacterized protein n=1 Tax=Zea ma... 72 4e-11
UniRef100_B3M6C2 GF24314 n=1 Tax=Drosophila ananassae RepID=B3M6... 72 4e-11
UniRef100_B8NQ79 GARP complex component (Vps54), putative n=1 Ta... 72 4e-11
UniRef100_B0CS25 Vacuolar protein sorting-associated protein 54 ... 72 4e-11
UniRef100_O10341 Uncharacterized 29.3 kDa protein n=1 Tax=Orgyia... 72 4e-11
UniRef100_Q9SI74 F23N19.12 n=1 Tax=Arabidopsis thaliana RepID=Q9... 71 5e-11
UniRef100_A9P9Z1 Predicted protein n=1 Tax=Populus trichocarpa R... 71 5e-11
UniRef100_Q2UAJ1 Vacuolar sorting protein VPS45 n=1 Tax=Aspergil... 71 5e-11
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 71 5e-11
UniRef100_UPI0001A2D5C6 UPI0001A2D5C6 related cluster n=1 Tax=Da... 71 7e-11
UniRef100_B3DJK6 Zgc:195037 protein n=1 Tax=Danio rerio RepID=B3... 71 7e-11
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 71 7e-11
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 71 7e-11
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 71 7e-11
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 71 7e-11
UniRef100_Q4PAL8 Putative uncharacterized protein n=1 Tax=Ustila... 71 7e-11
UniRef100_C5FBL2 Putative uncharacterized protein n=1 Tax=Micros... 71 7e-11
UniRef100_Q1HH32 Putative uncharacterized protein n=1 Tax=Anther... 70 9e-11
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 70 9e-11
UniRef100_B6KAY2 Putative uncharacterized protein n=2 Tax=Toxopl... 70 9e-11
UniRef100_C9JMC6 Putative uncharacterized protein ENSP0000038976... 70 9e-11
UniRef100_C9SUV5 Predicted protein n=1 Tax=Verticillium albo-atr... 70 9e-11
UniRef100_B6QUR5 Putative uncharacterized protein n=1 Tax=Penici... 70 9e-11
UniRef100_UPI0001B576F5 hypothetical protein StreC_09513 n=1 Tax... 70 1e-10
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 70 1e-10
UniRef100_Q3JTV6 Putative uncharacterized protein n=1 Tax=Burkho... 70 1e-10
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 70 1e-10
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 70 1e-10
UniRef100_B9HLT3 Predicted protein (Fragment) n=1 Tax=Populus tr... 70 1e-10
UniRef100_UPI0000DD9486 Os08g0280200 n=1 Tax=Oryza sativa Japoni... 70 1e-10
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 70 1e-10
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 70 1e-10
UniRef100_B9S5R2 Actin binding protein, putative n=1 Tax=Ricinus... 70 1e-10
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 70 1e-10
UniRef100_Q17L38 Vacuolar protein sorting (Fragment) n=1 Tax=Aed... 70 1e-10
UniRef100_Q6ZCX3 Formin-like protein 6 n=1 Tax=Oryza sativa Japo... 70 1e-10
UniRef100_Q6QXJ8 ORF55 n=1 Tax=Agrotis segetum granulovirus RepI... 69 2e-10
UniRef100_Q9SX31 F24J5.8 protein n=1 Tax=Arabidopsis thaliana Re... 69 2e-10
UniRef100_C5XK46 Putative uncharacterized protein Sb03g034710 n=... 69 2e-10
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 69 2e-10
UniRef100_C9JS29 Putative uncharacterized protein TNRC18 n=1 Tax... 69 2e-10
UniRef100_A8MSW5 Putative uncharacterized protein TNRC18 n=1 Tax... 69 2e-10
UniRef100_B8NIL1 Proline-rich, actin-associated protein Vrp1, pu... 69 2e-10
UniRef100_O15417-2 Isoform 2 of Trinucleotide repeat-containing ... 69 2e-10
UniRef100_P14918 Extensin n=1 Tax=Zea mays RepID=EXTN_MAIZE 69 2e-10
UniRef100_Q7XUL1 Os04g0498700 protein n=1 Tax=Oryza sativa Japon... 69 3e-10
UniRef100_Q9P944 Kexin-like protease KEX1 n=1 Tax=Pneumocystis m... 69 3e-10
UniRef100_B1B5Z7 Kexin-like protease KEX1 (Fragment) n=1 Tax=Pne... 69 3e-10
UniRef100_UPI0000E59252 mucin 2 precursor n=1 Tax=Homo sapiens R... 69 3e-10
UniRef100_Q91GH4 Putative uncharacterized protein n=1 Tax=Epiphy... 69 3e-10
UniRef100_Q9ASK4 Os01g0110500 protein n=1 Tax=Oryza sativa Japon... 69 3e-10
UniRef100_Q94JZ6 At3g24600 n=1 Tax=Arabidopsis thaliana RepID=Q9... 69 3e-10
UniRef100_B8AG26 Putative uncharacterized protein n=1 Tax=Oryza ... 69 3e-10
UniRef100_B8ACV5 Putative uncharacterized protein n=1 Tax=Oryza ... 69 3e-10
UniRef100_A8I1E4 Hydroxyproline-rich cell wall protein n=1 Tax=C... 69 3e-10
UniRef100_Q02817 Mucin-2 n=1 Tax=Homo sapiens RepID=MUC2_HUMAN 69 3e-10
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 68 4e-10
UniRef100_B1HHT1 Putative uncharacterized protein n=1 Tax=Burkho... 68 4e-10
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 68 4e-10
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 68 4e-10
UniRef100_C0Z3D3 AT2G14890 protein n=1 Tax=Arabidopsis thaliana ... 68 4e-10
UniRef100_B6U4I2 Putative uncharacterized protein n=1 Tax=Zea ma... 68 4e-10
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 68 4e-10
UniRef100_A4S1A8 Predicted protein n=1 Tax=Ostreococcus lucimari... 68 4e-10
UniRef100_Q95UQ2 Subtilisin-like protein n=1 Tax=Toxoplasma gond... 68 4e-10
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 68 6e-10
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 68 6e-10
UniRef100_Q9LV48 Protein kinase-like protein n=1 Tax=Arabidopsis... 68 6e-10
UniRef100_Q6QJ28 Putative uncharacterized protein (Fragment) n=1... 68 6e-10
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 68 6e-10
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 68 6e-10
UniRef100_C4J5K1 Putative uncharacterized protein n=1 Tax=Zea ma... 68 6e-10
UniRef100_B9H3S0 Predicted protein (Fragment) n=1 Tax=Populus tr... 68 6e-10
UniRef100_B6UDN6 Putative uncharacterized protein n=1 Tax=Zea ma... 68 6e-10
UniRef100_A8JKC9 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 68 6e-10
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 68 6e-10
UniRef100_A4RYQ1 Predicted protein n=1 Tax=Ostreococcus lucimari... 68 6e-10
UniRef100_A7SXP8 Predicted protein n=1 Tax=Nematostella vectensi... 68 6e-10
UniRef100_Q6CCD3 YALI0C10450p n=1 Tax=Yarrowia lipolytica RepID=... 68 6e-10
UniRef100_C9SV27 Pyruvate dehydrogenase protein X component n=1 ... 68 6e-10
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 68 6e-10
UniRef100_A8NHF3 Putative uncharacterized protein n=1 Tax=Coprin... 68 6e-10
UniRef100_Q18DZ3 Probable cell surface glycoprotein n=1 Tax=Halo... 68 6e-10
UniRef100_UPI0001982977 PREDICTED: hypothetical protein n=1 Tax=... 67 7e-10
UniRef100_UPI0000E1F407 PREDICTED: vacuolar protein sorting 54 i... 67 7e-10
UniRef100_UPI0000D9D459 PREDICTED: similar to vacuolar protein s... 67 7e-10
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 67 7e-10
UniRef100_Q9LT74 Similarity to late embryogenesis abundant prote... 67 7e-10
UniRef100_Q40385 Pistil extensin-like protein n=1 Tax=Nicotiana ... 67 7e-10
UniRef100_C1E833 Predicted protein n=1 Tax=Micromonas sp. RCC299... 67 7e-10
UniRef100_B7ZXA1 Putative uncharacterized protein n=1 Tax=Zea ma... 67 7e-10
UniRef100_A1IGC6 PERK1-like protein kinase n=1 Tax=Nicotiana tab... 67 7e-10
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 67 7e-10
UniRef100_Q5AD60 Putative uncharacterized protein LUV1 n=1 Tax=C... 64 9e-10
UniRef100_C4YJ79 Putative uncharacterized protein n=1 Tax=Candid... 64 9e-10
UniRef100_UPI00005DC1BB PERK10 (PROLINE-RICH EXTENSIN-LIKE RECEP... 67 1e-09
UniRef100_Q9FPQ5 Gamete-specific hydroxyproline-rich glycoprotei... 67 1e-09
UniRef100_Q9FFW5 Similarity to protein kinase n=1 Tax=Arabidopsi... 67 1e-09
UniRef100_Q9C660 Pto kinase interactor, putative n=1 Tax=Arabido... 67 1e-09
UniRef100_Q8S9B5 Matrix metalloproteinase n=1 Tax=Volvox carteri... 67 1e-09
UniRef100_C0Z219 AT2G14890 protein n=1 Tax=Arabidopsis thaliana ... 67 1e-09
UniRef100_B6SXS9 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 67 1e-09
UniRef100_B9QFG7 Subtilisin-like protein TgSUB1, putative n=1 Ta... 67 1e-09
UniRef100_B0WC20 Vacuolar protein sorting n=1 Tax=Culex quinquef... 67 1e-09
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 67 1e-09
UniRef100_Q8B9G6 Putative uncharacterized protein n=1 Tax=Rachip... 67 1e-09
UniRef100_Q9RCX9 Putative secreted proline-rich protein n=1 Tax=... 67 1e-09
UniRef100_Q9ARH1 Receptor protein kinase PERK1 n=1 Tax=Brassica ... 67 1e-09
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 67 1e-09
UniRef100_B9FLI1 Putative uncharacterized protein n=1 Tax=Oryza ... 67 1e-09
UniRef100_A9TWI4 Predicted protein n=1 Tax=Physcomitrella patens... 67 1e-09
UniRef100_A2Y786 Putative uncharacterized protein n=1 Tax=Oryza ... 67 1e-09
UniRef100_Q4CQ84 Cellulosomal scaffoldin anchoring protein, puta... 67 1e-09
UniRef100_B3MVK1 GF23569 n=1 Tax=Drosophila ananassae RepID=B3MV... 67 1e-09
UniRef100_Q5KJA8 Retrograde transport, endosome to Golgi-related... 67 1e-09
UniRef100_Q55WZ1 Putative uncharacterized protein n=1 Tax=Filoba... 67 1e-09
UniRef100_UPI0001A7B3C7 unknown protein n=1 Tax=Arabidopsis thal... 66 2e-09
UniRef100_UPI0001A7B0C1 protein binding / structural constituent... 66 2e-09
UniRef100_UPI0001982E1E PREDICTED: hypothetical protein n=1 Tax=... 66 2e-09
UniRef100_UPI000069F8DC UPI000069F8DC related cluster n=1 Tax=Xe... 66 2e-09
UniRef100_B8HBY3 Peptidase M23 n=1 Tax=Arthrobacter chlorophenol... 66 2e-09
UniRef100_B4BGA9 S-layer domain protein n=1 Tax=Clostridium ther... 66 2e-09
UniRef100_Q9SN46 Extensin-like protein n=1 Tax=Arabidopsis thali... 66 2e-09
UniRef100_C7IZA4 Os02g0162200 protein n=1 Tax=Oryza sativa Japon... 66 2e-09
UniRef100_C5Y027 Putative uncharacterized protein Sb04g010730 n=... 66 2e-09
UniRef100_B8AI70 Putative uncharacterized protein n=1 Tax=Oryza ... 66 2e-09
UniRef100_A8J1D8 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 66 2e-09
UniRef100_B9PU53 Subtilase family protein n=1 Tax=Toxoplasma gon... 66 2e-09
UniRef100_C6H7S2 Putative uncharacterized protein n=1 Tax=Ajello... 66 2e-09
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 66 2e-09
UniRef100_A5YT41 Probable cell surface glycoprotein n=1 Tax=uncu... 66 2e-09
UniRef100_UPI0001B54FB4 hypothetical protein StAA4_06026 n=1 Tax... 66 2e-09
UniRef100_UPI00017390C7 AGP19 (ARABINOGALACTAN-PROTEIN 19) n=1 T... 66 2e-09
UniRef100_UPI000151B182 hypothetical protein PGUG_04930 n=1 Tax=... 66 2e-09
UniRef100_UPI0000DD95B9 Os08g0528700 n=1 Tax=Oryza sativa Japoni... 66 2e-09
UniRef100_Q89392 A57R protein n=1 Tax=Paramecium bursaria Chlore... 66 2e-09
UniRef100_C9Z517 Putative secreted proline-rich protein n=1 Tax=... 66 2e-09
UniRef100_Q013M1 Chromosome 08 contig 1, DNA sequence n=1 Tax=Os... 66 2e-09
UniRef100_Q5CLM1 Putative uncharacterized protein n=1 Tax=Crypto... 66 2e-09
UniRef100_A3LN20 Predicted protein n=1 Tax=Pichia stipitis RepID... 66 2e-09
UniRef100_Q9S740 Lysine-rich arabinogalactan protein 19 n=1 Tax=... 66 2e-09
UniRef100_UPI0000DF06C6 Os02g0456000 n=1 Tax=Oryza sativa Japoni... 65 3e-09
UniRef100_Q825Z2 Putative proline-rich protein n=1 Tax=Streptomy... 65 3e-09
UniRef100_A7H9N7 Heavy metal translocating P-type ATPase n=1 Tax... 65 3e-09
UniRef100_C5NKF6 Intracellular motility protein A n=1 Tax=Burkho... 65 3e-09
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 65 3e-09
UniRef100_Q5VR46 Os01g0180000 protein n=1 Tax=Oryza sativa Japon... 65 3e-09
UniRef100_Q3EC03 Uncharacterized protein At2g14890.2 n=1 Tax=Ara... 65 3e-09
UniRef100_C5XMP4 Putative uncharacterized protein Sb03g003730 n=... 65 3e-09
UniRef100_C1EFP7 Receptor-like cell wall protein n=1 Tax=Micromo... 65 3e-09
UniRef100_B8ADK4 Putative uncharacterized protein n=1 Tax=Oryza ... 65 3e-09
UniRef100_B6SVB0 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 65 3e-09
UniRef100_A9RMD2 Predicted protein n=1 Tax=Physcomitrella patens... 65 3e-09
UniRef100_Q8MQG9 Prion-like-(Q/n-rich)-domain-bearing protein pr... 65 3e-09
UniRef100_O02123 Prion-like-(Q/n-rich)-domain-bearing protein pr... 65 3e-09
UniRef100_B6AGG8 Putative uncharacterized protein n=1 Tax=Crypto... 65 3e-09
UniRef100_Q6CEC1 YALI0B16822p n=1 Tax=Yarrowia lipolytica RepID=... 65 3e-09
UniRef100_C5M613 Putative uncharacterized protein n=1 Tax=Candid... 65 3e-09
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 65 3e-09
UniRef100_Q9C5S0 Classical arabinogalactan protein 9 n=1 Tax=Ara... 65 3e-09
UniRef100_UPI0000DB7674 PREDICTED: hypothetical protein n=1 Tax=... 65 4e-09
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 65 4e-09
UniRef100_A8C635 Putative uncharacterized protein n=1 Tax=Anther... 65 4e-09
UniRef100_A3QTV8 ORF149 n=1 Tax=Cyprinid herpesvirus 3 RepID=A3Q... 65 4e-09
UniRef100_A3QMW4 Putative uncharacterized protein n=1 Tax=Cyprin... 65 4e-09
UniRef100_Q73TB8 Putative uncharacterized protein n=1 Tax=Mycoba... 65 4e-09
UniRef100_Q2JMC8 TonB family protein n=1 Tax=Synechococcus sp. J... 65 4e-09
UniRef100_B1YPT1 Type VI secretion system Vgr family protein n=1... 65 4e-09
UniRef100_C4B111 Intracellular motility protein A n=4 Tax=Burkho... 65 4e-09
UniRef100_Q5ZB68 Os01g0594300 protein n=1 Tax=Oryza sativa Japon... 65 4e-09
UniRef100_O22016 HepB protein n=1 Tax=Cylindrotheca fusiformis R... 65 4e-09
UniRef100_C1ECP1 Resistance-nodulation-cell division superfamily... 65 4e-09
UniRef100_B9SHA4 ATP binding protein, putative n=1 Tax=Ricinus c... 65 4e-09
UniRef100_B9HGL7 Predicted protein n=1 Tax=Populus trichocarpa R... 65 4e-09
UniRef100_B9GFM1 Predicted protein n=1 Tax=Populus trichocarpa R... 65 4e-09
UniRef100_B8B912 Putative uncharacterized protein n=1 Tax=Oryza ... 65 4e-09
UniRef100_A9SIY2 Predicted protein n=1 Tax=Physcomitrella patens... 65 4e-09
UniRef100_A9PCL2 Putative uncharacterized protein n=1 Tax=Populu... 65 4e-09
UniRef100_B4MA01 GJ15821 n=1 Tax=Drosophila virilis RepID=B4MA01... 65 4e-09
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 65 5e-09
UniRef100_UPI0000E125D3 Os05g0552600 n=1 Tax=Oryza sativa Japoni... 65 5e-09
UniRef100_UPI0000223553 Hypothetical protein CBG15933 n=1 Tax=Ca... 65 5e-09
UniRef100_UPI0001A2C524 UPI0001A2C524 related cluster n=1 Tax=Da... 65 5e-09
UniRef100_UPI0001AE6B68 Mucin-2 precursor (Intestinal mucin-2). ... 65 5e-09
UniRef100_UPI00006C10F2 UPI00006C10F2 related cluster n=1 Tax=Ho... 65 5e-09
UniRef100_Q8VKN7 Putative uncharacterized protein n=1 Tax=Mycoba... 65 5e-09
UniRef100_Q9LIE8 Similarity to cell wall-plasma membrane linker ... 65 5e-09
UniRef100_Q7Q4D8 AGAP008353-PA (Fragment) n=1 Tax=Anopheles gamb... 65 5e-09
UniRef100_B9PLD5 Putative uncharacterized protein n=1 Tax=Toxopl... 65 5e-09
UniRef100_C9JDS7 Putative uncharacterized protein ENSP0000039622... 65 5e-09
UniRef100_Q7SCZ7 WASP-interacting protein-like protein vrp1p n=1... 65 5e-09
UniRef100_C5JBY8 Putative uncharacterized protein n=1 Tax=Ajello... 65 5e-09
UniRef100_A5DNS9 Putative uncharacterized protein n=1 Tax=Pichia... 65 5e-09
UniRef100_UPI0000E25181 PREDICTED: myeloid/lymphoid or mixed-lin... 64 6e-09
UniRef100_UPI0000DD9D0C Os11g0657400 n=1 Tax=Oryza sativa Japoni... 64 6e-09
UniRef100_A4VK92 TonB protein, C-terminal domain n=1 Tax=Pseudom... 64 6e-09
UniRef100_C7HD87 Type 3a cellulose-binding domain protein (Fragm... 64 6e-09
UniRef100_B9G8N7 Putative uncharacterized protein n=1 Tax=Oryza ... 64 6e-09
UniRef100_B9FLY6 Putative uncharacterized protein n=1 Tax=Oryza ... 64 6e-09
UniRef100_B8AWI6 Putative uncharacterized protein n=1 Tax=Oryza ... 64 6e-09
UniRef100_B6TRM4 Putative uncharacterized protein n=1 Tax=Zea ma... 64 6e-09
UniRef100_A9STR8 Predicted protein (Fragment) n=1 Tax=Physcomitr... 64 6e-09
UniRef100_A8IZG7 Plus agglutinin protein (Fragment) n=1 Tax=Chla... 64 6e-09
UniRef100_A4SBI2 Predicted protein n=1 Tax=Ostreococcus lucimari... 64 6e-09
UniRef100_B5SNN9 Homeobox A3 isoform a (Predicted) n=1 Tax=Otole... 64 6e-09
UniRef100_B6KI42 Subtilase family serine protease, putative n=1 ... 64 6e-09
UniRef100_B4IMI5 GM16182 n=1 Tax=Drosophila sechellia RepID=B4IM... 64 6e-09
UniRef100_A8MVY7 Putative uncharacterized protein ENSP0000038013... 64 6e-09
UniRef100_A8N135 Predicted protein n=1 Tax=Coprinopsis cinerea o... 64 6e-09
UniRef100_Q9UMN6-2 Isoform 2 of Histone-lysine N-methyltransfera... 64 6e-09
UniRef100_Q9UMN6 Histone-lysine N-methyltransferase MLL4 n=1 Tax... 64 6e-09
UniRef100_UPI00019272F7 PREDICTED: similar to scattered CG3766-P... 64 8e-09
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 64 8e-09
UniRef100_UPI0000D9EBB1 PREDICTED: myeloid/lymphoid or mixed-lin... 64 8e-09
UniRef100_UPI0001A2D6D8 UPI0001A2D6D8 related cluster n=1 Tax=Da... 64 8e-09
UniRef100_UPI00017B3BB9 UPI00017B3BB9 related cluster n=1 Tax=Te... 64 8e-09
UniRef100_UPI00017B3BB8 UPI00017B3BB8 related cluster n=1 Tax=Te... 64 8e-09
UniRef100_A3DIC9 S-layer-like domain containing protein n=1 Tax=... 64 8e-09
UniRef100_B4V800 Secreted protein n=1 Tax=Streptomyces sp. Mg1 R... 64 8e-09
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 64 8e-09
UniRef100_Q43558 Proline rich protein n=1 Tax=Medicago sativa Re... 64 8e-09
UniRef100_Q09084 Extensin (Class II) n=1 Tax=Solanum lycopersicu... 64 8e-09
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 64 8e-09
UniRef100_B9HIU4 Predicted protein (Fragment) n=1 Tax=Populus tr... 64 8e-09
UniRef100_B4FC70 Putative uncharacterized protein n=1 Tax=Zea ma... 64 8e-09
UniRef100_A3BFF7 Putative uncharacterized protein n=1 Tax=Oryza ... 64 8e-09
UniRef100_B9PHB2 Putative uncharacterized protein n=1 Tax=Toxopl... 64 8e-09
UniRef100_B3NC39 GG15230 n=1 Tax=Drosophila erecta RepID=B3NC39_... 64 8e-09
UniRef100_C5G9C0 Putative uncharacterized protein n=1 Tax=Ajello... 64 8e-09
UniRef100_A8N9F0 Predicted protein n=1 Tax=Coprinopsis cinerea o... 64 8e-09
UniRef100_UPI000192D0B3 cpf1 genetically-interacting protein 1, ... 64 1e-08
UniRef100_UPI0001925A99 PREDICTED: hypothetical protein, partial... 64 1e-08
UniRef100_Q69ZN8 MKIAA1205 protein (Fragment) n=3 Tax=Mus muscul... 64 1e-08
UniRef100_UPI000017E0D6 PREDICTED: similar to Homeobox protein H... 64 1e-08
UniRef100_UPI00016E59B7 UPI00016E59B7 related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E59B6 UPI00016E59B6 related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E599B UPI00016E599B related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E599A UPI00016E599A related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E5999 UPI00016E5999 related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E597F UPI00016E597F related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_UPI00016E595C UPI00016E595C related cluster n=1 Tax=Ta... 64 1e-08
UniRef100_C5BID0 Putative uncharacterized protein n=1 Tax=Teredi... 64 1e-08
UniRef100_A4X4V1 Putative uncharacterized protein n=1 Tax=Salini... 64 1e-08
UniRef100_Q9SPM0 Extensin-like protein n=1 Tax=Zea mays RepID=Q9... 64 1e-08
UniRef100_Q41814 Hydroxyproline-rich glycoprotein n=1 Tax=Zea ma... 64 1e-08
UniRef100_Q3HTK2 Pherophorin-C5 protein n=1 Tax=Chlamydomonas re... 64 1e-08
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 64 1e-08
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 64 1e-08
UniRef100_A9PGI2 Putative uncharacterized protein n=1 Tax=Populu... 64 1e-08
UniRef100_A9P8J4 Predicted protein n=1 Tax=Populus trichocarpa R... 64 1e-08
UniRef100_A8J9H7 Mastigoneme-like flagellar protein n=1 Tax=Chla... 64 1e-08
UniRef100_A4S9A6 Predicted protein n=1 Tax=Ostreococcus lucimari... 64 1e-08
UniRef100_A2XD25 Putative uncharacterized protein n=1 Tax=Oryza ... 64 1e-08
UniRef100_C3Z6S3 Putative uncharacterized protein n=1 Tax=Branch... 64 1e-08
UniRef100_B4IQK0 GM19288 n=1 Tax=Drosophila sechellia RepID=B4IQ... 64 1e-08
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 64 1e-08
UniRef100_P41479 Uncharacterized 24.1 kDa protein in LEF4-P33 in... 64 1e-08
UniRef100_P02831 Homeobox protein Hox-A3 n=1 Tax=Mus musculus Re... 64 1e-08
UniRef100_UPI000198414B PREDICTED: hypothetical protein isoform ... 63 1e-08
UniRef100_UPI000198414A PREDICTED: hypothetical protein isoform ... 63 1e-08
UniRef100_UPI0000DA2692 PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_Q4A373 Putative lectin protein n=2 Tax=Emiliania huxle... 63 1e-08
UniRef100_Q0GYD2 Putative uncharacterized protein n=1 Tax=Plutel... 63 1e-08
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 63 1e-08
UniRef100_B4YB56 BimA n=1 Tax=Burkholderia pseudomallei RepID=B4... 63 1e-08
UniRef100_Q8W158 Anther-specific proline-rich protein n=1 Tax=Br... 63 1e-08
UniRef100_Q852P0 Pherophorin n=1 Tax=Volvox carteri f. nagariens... 63 1e-08
UniRef100_Q41719 Hydroxyproline-rich glycoprotein n=1 Tax=Zea di... 63 1e-08
UniRef100_Q39789 Proline-rich cell wall protein n=1 Tax=Gossypiu... 63 1e-08
UniRef100_Q39620 VSP-3 protein n=1 Tax=Chlamydomonas reinhardtii... 63 1e-08
UniRef100_O65760 Extensin (Fragment) n=1 Tax=Cicer arietinum Rep... 63 1e-08
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 63 1e-08
UniRef100_B9RPP5 Early nodulin 20, putative n=1 Tax=Ricinus comm... 63 1e-08
UniRef100_A9T9L1 Predicted protein n=1 Tax=Physcomitrella patens... 63 1e-08
UniRef100_A8J790 Hydroxyproline-rich glycoprotein, stress-induce... 63 1e-08
UniRef100_Q29PH7 GA17672 n=1 Tax=Drosophila pseudoobscura pseudo... 63 1e-08
UniRef100_C3ZT83 Putative uncharacterized protein n=1 Tax=Branch... 63 1e-08
UniRef100_B6KES5 Putative uncharacterized protein n=1 Tax=Toxopl... 63 1e-08
UniRef100_B5E0E8 GA24203 n=1 Tax=Drosophila pseudoobscura pseudo... 63 1e-08
UniRef100_B4PI78 GE20613 n=1 Tax=Drosophila yakuba RepID=B4PI78_... 63 1e-08
UniRef100_B4MMR0 GK17563 n=1 Tax=Drosophila willistoni RepID=B4M... 63 1e-08
UniRef100_B4GKS6 GL26149 n=1 Tax=Drosophila persimilis RepID=B4G... 63 1e-08
UniRef100_A2GVH9 Putative uncharacterized protein (Fragment) n=1... 63 1e-08
UniRef100_Q5KGJ5 Putative uncharacterized protein n=1 Tax=Filoba... 63 1e-08
UniRef100_C5GNH0 Putative uncharacterized protein n=1 Tax=Ajello... 63 1e-08
UniRef100_Q6IN36 WAS/WASL-interacting protein family member 1 n=... 63 1e-08
UniRef100_Q03211 Pistil-specific extensin-like protein n=1 Tax=N... 63 1e-08
UniRef100_UPI0001B4102B Rhs element Vgr protein n=1 Tax=Burkhold... 63 2e-08
UniRef100_UPI000198316D PREDICTED: hypothetical protein n=1 Tax=... 63 2e-08
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 63 2e-08
UniRef100_UPI000179680F PREDICTED: similar to WW domain-binding ... 63 2e-08
UniRef100_UPI00016B1084 Rhs element Vgr protein n=1 Tax=Burkhold... 63 2e-08
UniRef100_UPI00005A37C2 PREDICTED: similar to Mucin-5B precursor... 63 2e-08
UniRef100_UPI0000EB4AD1 UPI0000EB4AD1 related cluster n=1 Tax=Ca... 63 2e-08
UniRef100_A8DJ06 UL36 (Fragment) n=1 Tax=Gallid herpesvirus 2 Re... 63 2e-08
UniRef100_Q2T918 Rhs element Vgr protein n=1 Tax=Burkholderia th... 63 2e-08
UniRef100_A7NK84 Putative uncharacterized protein n=1 Tax=Roseif... 63 2e-08
UniRef100_A7HFY4 ABC transporter related n=1 Tax=Anaeromyxobacte... 63 2e-08
UniRef100_C7Q7I9 Serine/threonine protein kinase n=1 Tax=Catenul... 63 2e-08
UniRef100_Q6QJ26 Putative uncharacterized protein (Fragment) n=1... 63 2e-08
UniRef100_Q69L88 cDNA clone:J013069I08, full insert sequence n=1... 63 2e-08
UniRef100_Q4KXD9 Early salt stress and cold acclimation-induced ... 63 2e-08
UniRef100_Q39763 Proline-rich cell wall protein n=1 Tax=Gossypiu... 63 2e-08
UniRef100_Q00TR5 Homology to unknown gene n=1 Tax=Ostreococcus t... 63 2e-08
UniRef100_O65530 Putative uncharacterized protein AT4g32710 n=1 ... 63 2e-08
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 63 2e-08
UniRef100_A9U3I4 Predicted protein n=1 Tax=Physcomitrella patens... 63 2e-08
UniRef100_A7PLL6 Chromosome chr7 scaffold_20, whole genome shotg... 63 2e-08
UniRef100_B4NPA2 GK15614 n=1 Tax=Drosophila willistoni RepID=B4N... 63 2e-08
UniRef100_Q7SFQ1 Predicted protein n=1 Tax=Neurospora crassa Rep... 63 2e-08
UniRef100_P93329 Early nodulin-20 n=1 Tax=Medicago truncatula Re... 63 2e-08
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI000198409E PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI0001982858 PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI00015B5C8A PREDICTED: similar to formin 1,2/cappucc... 62 2e-08
UniRef100_UPI000155BFF2 PREDICTED: hypothetical protein, partial... 62 2e-08
UniRef100_UPI000162C860 nascent polypeptide-associated complex a... 62 2e-08
UniRef100_UPI00016E7DED UPI00016E7DED related cluster n=1 Tax=Ta... 62 2e-08
UniRef100_Q6X248 UL36 very large tegument protein n=1 Tax=Bovine... 62 2e-08
UniRef100_A8DJ25 UL36 (Fragment) n=1 Tax=Gallid herpesvirus 2 Re... 62 2e-08
UniRef100_Q8LK02 Putative uncharacterized protein S126P21.5 n=1 ... 62 2e-08
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 62 2e-08
UniRef100_O22514 Proline rich protein n=1 Tax=Santalum album Rep... 62 2e-08
UniRef100_C5YGC5 Putative uncharacterized protein Sb06g016390 n=... 62 2e-08
UniRef100_B6SUZ1 Putative uncharacterized protein n=1 Tax=Zea ma... 62 2e-08
UniRef100_A7XQ02 Latex protein n=1 Tax=Morus alba RepID=A7XQ02_M... 62 2e-08
UniRef100_A7QNX2 Chromosome chr1 scaffold_135, whole genome shot... 62 2e-08
UniRef100_B9QDW3 Putative uncharacterized protein n=1 Tax=Toxopl... 62 2e-08
UniRef100_B6KLQ3 Putative uncharacterized protein n=1 Tax=Toxopl... 62 2e-08
UniRef100_B4HU68 GM13974 n=1 Tax=Drosophila sechellia RepID=B4HU... 62 2e-08
UniRef100_A8Y2E4 Putative uncharacterized protein (Fragment) n=1... 62 2e-08
UniRef100_Q6FSJ1 Similarities with uniprot|P47179 Saccharomyces ... 62 2e-08
UniRef100_A5E068 Putative uncharacterized protein n=1 Tax=Lodder... 62 2e-08
UniRef100_UPI00019829F7 PREDICTED: hypothetical protein n=1 Tax=... 62 3e-08
UniRef100_UPI00003BD061 hypothetical protein DEHA0A01639g n=1 Ta... 62 3e-08
UniRef100_UPI00016E98C2 UPI00016E98C2 related cluster n=1 Tax=Ta... 62 3e-08
UniRef100_Q7ZYY2 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 62 3e-08
UniRef100_Q6NYX7 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 62 3e-08
UniRef100_Q5RGR6 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 62 3e-08
UniRef100_Q8UZB4 Putative uncharacterized protein n=1 Tax=Grapev... 62 3e-08
UniRef100_Q7U3X4 Putative uncharacterized protein n=1 Tax=Synech... 62 3e-08
UniRef100_Q2JQ30 Putative uncharacterized protein n=1 Tax=Synech... 62 3e-08
UniRef100_Q1D2V2 Adventurous gliding motility protein AgmX n=1 T... 62 3e-08
UniRef100_Q0RSN4 Putative uncharacterized protein n=1 Tax=Franki... 62 3e-08
UniRef100_A7NR83 Putative uncharacterized protein n=1 Tax=Roseif... 62 3e-08
UniRef100_A5US08 Conserved repeat domain n=1 Tax=Roseiflexus sp.... 62 3e-08
UniRef100_Q84BD5 Adventurous gliding motility protein X n=1 Tax=... 62 3e-08