[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= DC597039 MPD058a12_r
(306 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
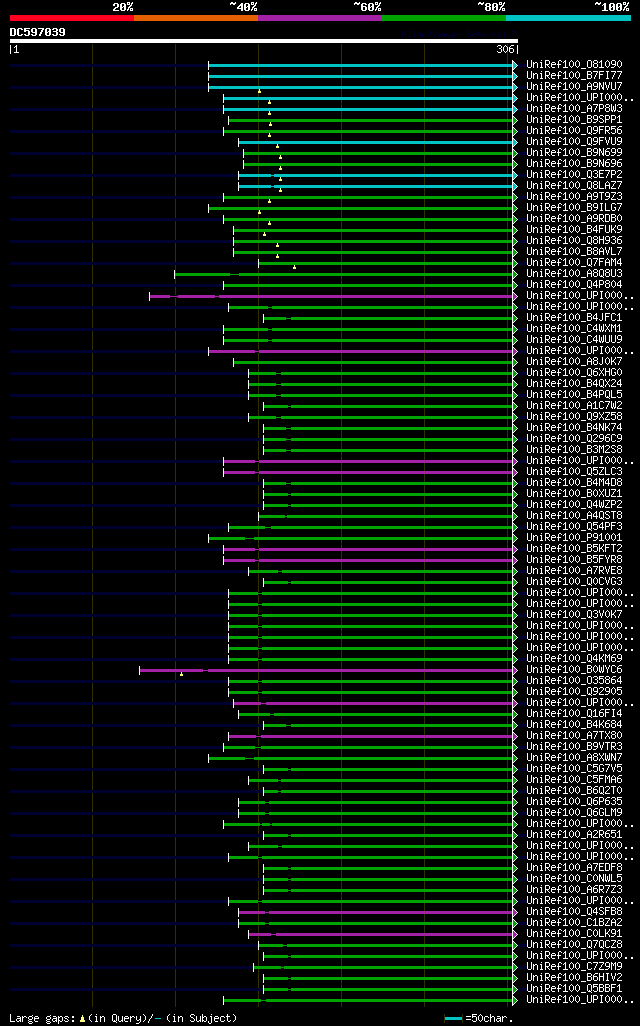
significant alignments:[graphical|details]
UniRef100_O81090 Putative JUN kinase activation domain binding p... 123 7e-27
UniRef100_B7FI77 Putative uncharacterized protein n=1 Tax=Medica... 123 7e-27
UniRef100_A9NVU7 Putative uncharacterized protein n=1 Tax=Picea ... 95 3e-18
UniRef100_UPI0001982DA0 PREDICTED: similar to JAB n=1 Tax=Vitis ... 94 4e-18
UniRef100_A7P8W3 Chromosome chr3 scaffold_8, whole genome shotgu... 94 4e-18
UniRef100_B9SPP1 Jun activation domain binding protein, putative... 91 3e-17
UniRef100_Q9FR56 JAB n=1 Tax=Solanum lycopersicum RepID=Q9FR56_S... 90 7e-17
UniRef100_Q9FVU9 COP9 signalosome complex subunit 5a n=1 Tax=Ara... 87 8e-16
UniRef100_B9N699 Predicted protein (Fragment) n=1 Tax=Populus tr... 85 2e-15
UniRef100_B9N696 Predicted protein n=1 Tax=Populus trichocarpa R... 85 2e-15
UniRef100_Q3E7P2 Putative uncharacterized protein At1g22920.2 n=... 82 2e-14
UniRef100_Q8LAZ7 COP9 signalosome complex subunit 5b n=2 Tax=Ara... 82 2e-14
UniRef100_A9T9Z3 Predicted protein n=1 Tax=Physcomitrella patens... 81 3e-14
UniRef100_B9ILG7 Predicted protein n=1 Tax=Populus trichocarpa R... 79 2e-13
UniRef100_A9RDB0 Predicted protein n=1 Tax=Physcomitrella patens... 77 5e-13
UniRef100_B4FUK9 COP9 signalosome complex subunit 5b n=1 Tax=Zea... 73 9e-12
UniRef100_Q8H936 Os04g0654700 protein n=3 Tax=Oryza sativa RepID... 72 1e-11
UniRef100_B8AVL7 Putative uncharacterized protein n=1 Tax=Oryza ... 72 1e-11
UniRef100_Q7FAM4 OSJNBa0071I13.2 protein n=2 Tax=Oryza sativa Ja... 67 6e-10
UniRef100_A8Q8U3 COP9 signalosome complex subunit 5, putative n=... 61 3e-08
UniRef100_Q4P804 COP9 signalosome complex subunit 5 n=1 Tax=Usti... 61 3e-08
UniRef100_UPI000180D2A5 PREDICTED: similar to COP9 constitutive ... 60 6e-08
UniRef100_UPI00015B5D89 PREDICTED: similar to jun activation dom... 60 8e-08
UniRef100_B4JFC1 GH19288 n=1 Tax=Drosophila grimshawi RepID=B4JF... 60 8e-08
UniRef100_C4WXM1 ACYPI006786 protein n=1 Tax=Acyrthosiphon pisum... 60 1e-07
UniRef100_C4WUU9 ACYPI006786 protein n=1 Tax=Acyrthosiphon pisum... 60 1e-07
UniRef100_UPI000051AA9B PREDICTED: similar to COP9 constitutive ... 59 1e-07
UniRef100_A8J0K7 COP signalosome subunit 5 (Fragment) n=1 Tax=Ch... 59 1e-07
UniRef100_Q6XHG0 Similar to Drosophila melanogaster CSN5 (Fragme... 59 2e-07
UniRef100_B4QX24 GD19103 n=1 Tax=Drosophila simulans RepID=B4QX2... 59 2e-07
UniRef100_B4PQL5 CSN5 n=2 Tax=melanogaster subgroup RepID=B4PQL5... 59 2e-07
UniRef100_A1C7W2 COP9 signalosome subunit CsnE n=1 Tax=Aspergill... 59 2e-07
UniRef100_Q9XZ58 COP9 signalosome complex subunit 5 n=2 Tax=mela... 59 2e-07
UniRef100_B4NK74 GK14483 n=1 Tax=Drosophila willistoni RepID=B4N... 59 2e-07
UniRef100_Q296C9 GA13321 n=2 Tax=pseudoobscura subgroup RepID=Q2... 59 2e-07
UniRef100_B3M2S8 GF18500 n=1 Tax=Drosophila ananassae RepID=B3M2... 59 2e-07
UniRef100_UPI0000ECD101 hypothetical protein LOC426579 n=1 Tax=G... 58 3e-07
UniRef100_Q5ZLC3 Putative uncharacterized protein n=1 Tax=Gallus... 58 3e-07
UniRef100_B4M4D8 GJ10255 n=1 Tax=Drosophila virilis RepID=B4M4D8... 58 4e-07
UniRef100_B0XUZ1 COP9 signalosome subunit 5 (CsnE), putative n=1... 58 4e-07
UniRef100_Q4WZP2 COP9 signalosome complex subunit 5 n=1 Tax=Aspe... 58 4e-07
UniRef100_A4QST8 Putative uncharacterized protein n=1 Tax=Magnap... 57 5e-07
UniRef100_Q54PF3 COP9 signalosome complex subunit 5 n=1 Tax=Dict... 57 5e-07
UniRef100_P91001 COP9 signalosome complex subunit 5 n=1 Tax=Caen... 57 5e-07
UniRef100_B5KFT2 Putative JUN activation binding protein variant... 57 6e-07
UniRef100_B5FYR8 Putative JUN activation binding protein variant... 57 6e-07
UniRef100_A7RVE8 Predicted protein n=1 Tax=Nematostella vectensi... 57 6e-07
UniRef100_Q0CVG3 COP9 signalosome complex subunit 5 n=1 Tax=Aspe... 57 6e-07
UniRef100_UPI000155ECA5 PREDICTED: similar to COP9 signalosome s... 56 1e-06
UniRef100_UPI00005A503F PREDICTED: similar to COP9 signalosome c... 56 1e-06
UniRef100_Q3V0K7 Putative uncharacterized protein n=2 Tax=Euther... 56 1e-06
UniRef100_UPI00004A6ED0 PREDICTED: similar to COP9 signalosome c... 56 1e-06
UniRef100_UPI00005BE1EA PREDICTED: similar to COP9 signalosome s... 56 1e-06
UniRef100_UPI00017C3CF7 PREDICTED: similar to COP9 signalosome s... 56 1e-06
UniRef100_Q4KM69 COP9 constitutive photomorphogenic homolog subu... 56 1e-06
UniRef100_B0WYC6 COP9 signalosome complex subunit 5 n=1 Tax=Cule... 56 1e-06
UniRef100_O35864 COP9 signalosome complex subunit 5 n=1 Tax=Mus ... 56 1e-06
UniRef100_Q92905 COP9 signalosome complex subunit 5 n=1 Tax=Homo... 56 1e-06
UniRef100_UPI000049323C PREDICTED: similar to COP9 constitutive ... 56 1e-06
UniRef100_Q16FI4 Jun activation domain binding protein n=1 Tax=A... 56 1e-06
UniRef100_B4K684 GI23503 n=1 Tax=Drosophila mojavensis RepID=B4K... 56 1e-06
UniRef100_A7TX80 COP9 constitutive photomorphogenic-like subunit... 55 2e-06
UniRef100_B9VTR3 JAB-MPN domain protein n=1 Tax=Bombyx mori RepI... 55 2e-06
UniRef100_A8XWN7 C. briggsae CBR-CSN-5 protein n=1 Tax=Caenorhab... 55 2e-06
UniRef100_C5G7V5 COP9 signalosome complex subunit 5 n=2 Tax=Ajel... 55 2e-06
UniRef100_C5FMA6 COP9 signalosome complex subunit 5 n=1 Tax=Micr... 55 2e-06
UniRef100_B6Q2T0 COP9 signalosome subunit CsnE n=1 Tax=Penicilli... 55 2e-06
UniRef100_Q6P635 COP9 signalosome complex subunit 5 n=1 Tax=Xeno... 55 2e-06
UniRef100_Q6GLM9 COP9 signalosome complex subunit 5 n=1 Tax=Xeno... 55 2e-06
UniRef100_UPI000186CB89 COP9 signalosome complex subunit, putati... 55 2e-06
UniRef100_A2R651 Contig An15c0220, complete genome n=1 Tax=Asper... 55 2e-06
UniRef100_UPI0001924726 PREDICTED: similar to predicted protein,... 55 3e-06
UniRef100_UPI00005E7EC3 PREDICTED: similar to COP9 constitutive ... 55 3e-06
UniRef100_A7EDF8 Putative uncharacterized protein n=1 Tax=Sclero... 55 3e-06
UniRef100_C0NWL5 COP9 signalosome complex subunit n=2 Tax=Ajello... 54 4e-06
UniRef100_A6R7Z3 COP9 signalosome complex subunit 5 n=1 Tax=Ajel... 54 4e-06
UniRef100_UPI0000EDE82A PREDICTED: similar to COP9 constitutive ... 54 5e-06
UniRef100_Q4SFB8 Chromosome 6 SCAF14605, whole genome shotgun se... 54 5e-06
UniRef100_C1BZA2 COP9 signalosome complex subunit 5 n=1 Tax=Esox... 54 5e-06
UniRef100_C0LK91 COP9 signalosome subunit 5 (Fragment) n=1 Tax=M... 54 5e-06
UniRef100_Q7QCZ8 AGAP002880-PA (Fragment) n=1 Tax=Anopheles gamb... 54 5e-06
UniRef100_UPI00002343F2 hypothetical protein AN2129.2 n=1 Tax=As... 54 7e-06
UniRef100_C7Z9M9 Predicted protein n=1 Tax=Nectria haematococca ... 54 7e-06
UniRef100_B6HIV2 Pc21g01770 protein n=1 Tax=Penicillium chrysoge... 54 7e-06
UniRef100_Q5BBF1 COP9 signalosome complex subunit 5 n=2 Tax=Emer... 54 7e-06
UniRef100_UPI0000D570F9 PREDICTED: similar to jun activation dom... 53 9e-06
UniRef100_C5PDV3 COP9 signalosome complex subunit 5, putative n=... 53 9e-06