[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= BP055769 SPDL060g10_f
(461 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
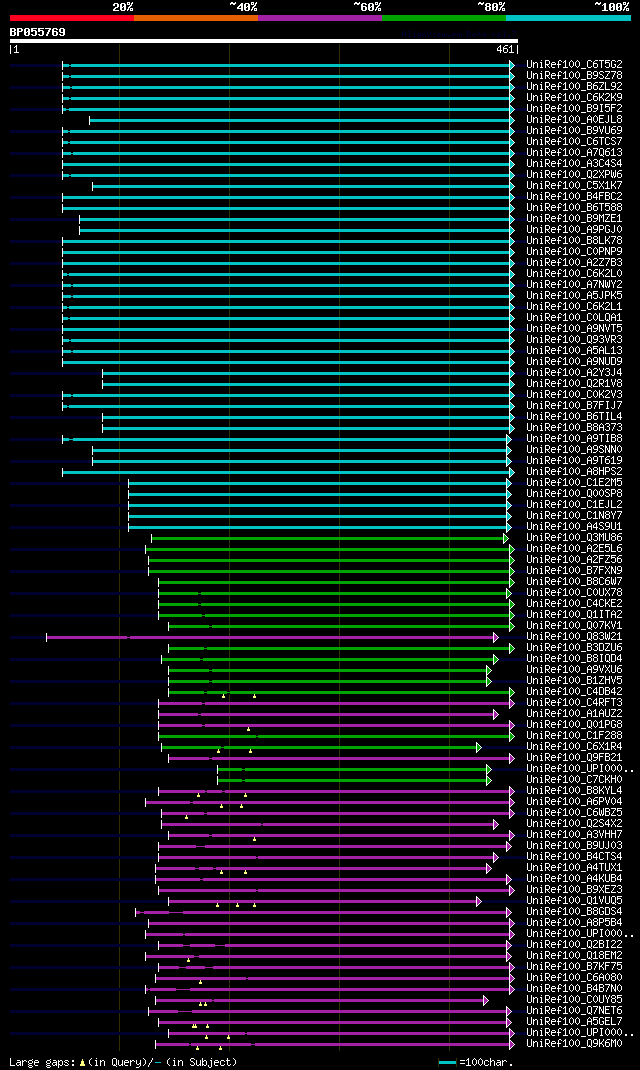
significant alignments:[graphical|details]
UniRef100_C6T5G2 Putative uncharacterized protein (Fragment) n=1... 262 8e-69
UniRef100_B9SZ78 Dtdp-glucose 4-6-dehydratase, putative n=1 Tax=... 260 4e-68
UniRef100_B6ZL92 GDP-D-mannose-3',5'-epimerase n=1 Tax=Prunus pe... 259 8e-68
UniRef100_C6K2K9 GDP-mannose 3',5'-epimerase n=1 Tax=Solanum lyc... 257 2e-67
UniRef100_B9I5F2 Predicted protein n=1 Tax=Populus trichocarpa R... 257 2e-67
UniRef100_A0EJL8 GDP-D-mannose-3',5'-epimerase n=1 Tax=Malpighia... 257 2e-67
UniRef100_B9VU69 GDP-mannose-3',5'-epimerase n=1 Tax=Caragana ko... 257 3e-67
UniRef100_C6TCS7 Putative uncharacterized protein n=1 Tax=Glycin... 256 5e-67
UniRef100_A7Q613 Chromosome chr14 scaffold_54, whole genome shot... 256 5e-67
UniRef100_A3C4S4 GDP-mannose 3,5-epimerase 1 n=2 Tax=Oryza sativ... 256 7e-67
UniRef100_Q2XPW6 NAD-dependent epimerase/dehydratase family prot... 255 9e-67
UniRef100_C5X1K7 Putative uncharacterized protein Sb01g021890 n=... 255 9e-67
UniRef100_B4FBC2 Putative uncharacterized protein n=1 Tax=Zea ma... 255 9e-67
UniRef100_B6T588 GDP-mannose 3,5-epimerase 1 n=1 Tax=Zea mays Re... 255 1e-66
UniRef100_B9MZE1 Predicted protein n=1 Tax=Populus trichocarpa R... 254 2e-66
UniRef100_A9PGJ0 Putative uncharacterized protein n=1 Tax=Populu... 254 2e-66
UniRef100_B8LK78 Putative uncharacterized protein n=1 Tax=Picea ... 254 3e-66
UniRef100_C0PNP9 Putative uncharacterized protein n=1 Tax=Zea ma... 253 4e-66
UniRef100_A2Z7B3 GDP-mannose 3,5-epimerase 1 n=1 Tax=Oryza sativ... 253 4e-66
UniRef100_C6K2L0 GDP-mannose 3',5'-epimerase n=1 Tax=Solanum lyc... 253 5e-66
UniRef100_A7NWY2 Chromosome chr5 scaffold_2, whole genome shotgu... 253 5e-66
UniRef100_A5JPK5 GDP-mannose-3',5'-epimerase n=1 Tax=Vitis vinif... 253 5e-66
UniRef100_C6K2L1 GDP-mannose 3',5'-epimerase n=1 Tax=Solanum pen... 251 1e-65
UniRef100_C0LQA1 GDP-D-mannose-3',5'-epimerase n=1 Tax=Malus x d... 251 2e-65
UniRef100_A9NVT5 Putative uncharacterized protein n=1 Tax=Picea ... 251 2e-65
UniRef100_Q93VR3 GDP-mannose 3,5-epimerase n=2 Tax=Arabidopsis t... 251 2e-65
UniRef100_A5AL13 Putative uncharacterized protein n=1 Tax=Vitis ... 250 3e-65
UniRef100_A9NUD9 Putative uncharacterized protein n=2 Tax=Picea ... 248 1e-64
UniRef100_A2Y3J4 Putative uncharacterized protein n=1 Tax=Oryza ... 248 1e-64
UniRef100_Q2R1V8 GDP-mannose 3,5-epimerase 2 n=2 Tax=Oryza sativ... 248 1e-64
UniRef100_C0K2V3 GDP-D-mannose-3',5'-epimerase n=1 Tax=Ribes nig... 248 2e-64
UniRef100_B7FIJ7 Putative uncharacterized protein n=1 Tax=Medica... 247 3e-64
UniRef100_B6TIL4 GDP-mannose 3,5-epimerase 2 n=1 Tax=Zea mays Re... 242 8e-63
UniRef100_B8A373 Putative uncharacterized protein n=1 Tax=Zea ma... 242 1e-62
UniRef100_A9TIB8 Predicted protein n=1 Tax=Physcomitrella patens... 232 8e-60
UniRef100_A9SNN0 Predicted protein n=1 Tax=Physcomitrella patens... 228 2e-58
UniRef100_A9T619 Predicted protein n=1 Tax=Physcomitrella patens... 227 3e-58
UniRef100_A8HPS2 Sugar nucleotide epimerase n=1 Tax=Chlamydomona... 207 2e-52
UniRef100_C1E2M5 Predicted protein n=1 Tax=Micromonas sp. RCC299... 186 7e-46
UniRef100_Q00SP8 dTDP-glucose 4-6-dehydratase/UDP-glucuronic aci... 185 2e-45
UniRef100_C1EJL2 Predicted protein n=1 Tax=Micromonas sp. RCC299... 183 6e-45
UniRef100_C1N8Y7 Predicted protein n=1 Tax=Micromonas pusilla CC... 181 2e-44
UniRef100_A4S9U1 Predicted protein n=1 Tax=Ostreococcus lucimari... 181 3e-44
UniRef100_Q3MU86 GDP-mannose-3'',5''-epimerase n=1 Tax=Oryza sat... 145 1e-33
UniRef100_A2E5L6 Epimerase/dehydratase, putative n=1 Tax=Trichom... 130 5e-29
UniRef100_A2FZ56 AT5g28840/F7P1_20, putative n=1 Tax=Trichomonas... 129 1e-28
UniRef100_B7FXN9 Nad-dependent epimerase/dehydratase n=1 Tax=Pha... 122 2e-26
UniRef100_B8C6W7 Dual function enzyme: UDP-glucose 4-epimerase n... 115 2e-24
UniRef100_C0UX78 Nucleoside-diphosphate-sugar epimerase n=1 Tax=... 113 7e-24
UniRef100_C4CKE2 Nucleoside-diphosphate-sugar epimerase n=1 Tax=... 110 5e-23
UniRef100_Q1ITA2 NAD-dependent epimerase/dehydratase n=1 Tax=Can... 107 3e-22
UniRef100_Q07KV1 NAD-dependent epimerase/dehydratase n=1 Tax=Rho... 96 1e-18
UniRef100_Q83W21 Ata17 protein n=1 Tax=Saccharothrix mutabilis s... 96 1e-18
UniRef100_B3DZU6 Nucleoside-diphosphate-sugar epimerase n=1 Tax=... 93 8e-18
UniRef100_B8IQD4 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 90 9e-17
UniRef100_A9VXU6 NAD-dependent epimerase/dehydratase n=2 Tax=Met... 89 2e-16
UniRef100_B1ZHV5 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 88 3e-16
UniRef100_C4DB42 Nucleoside-diphosphate-sugar epimerase n=1 Tax=... 82 2e-14
UniRef100_C4RFT3 Ata17 protein n=1 Tax=Micromonospora sp. ATCC 3... 80 5e-14
UniRef100_A1AUZ2 NAD-dependent epimerase/dehydratase n=1 Tax=Pel... 80 9e-14
UniRef100_Q01PG8 NAD-dependent epimerase/dehydratase n=1 Tax=Can... 76 1e-12
UniRef100_C1F288 NAD dependent epimerase/dehydratase family prot... 75 2e-12
UniRef100_C6X1R4 Sugar epimerase BlmG n=1 Tax=Flavobacteriaceae ... 75 3e-12
UniRef100_Q9FB21 Sugar epimerase BlmG n=1 Tax=Streptomyces verti... 72 1e-11
UniRef100_UPI0000383029 COG0451: Nucleoside-diphosphate-sugar ep... 70 6e-11
UniRef100_C7CKH0 GDP-mannose 3,5-epimerase n=2 Tax=Methylobacter... 70 6e-11
UniRef100_B8KYL4 GDP-mannose 3,5-epimerase 1 n=1 Tax=gamma prote... 70 6e-11
UniRef100_A6PV04 NAD-dependent epimerase/dehydratase n=1 Tax=Vic... 70 1e-10
UniRef100_C6WBZ5 NAD-dependent epimerase/dehydratase n=1 Tax=Act... 69 2e-10
UniRef100_Q2S4X2 Sugar epimerase BlmG n=1 Tax=Salinibacter ruber... 68 3e-10
UniRef100_A3VHH7 UDP-glucose 4-epimerase n=1 Tax=Rhodobacterales... 68 3e-10
UniRef100_B9UJ03 NAD-dependent sugar epimerase n=1 Tax=Streptomy... 67 5e-10
UniRef100_B4CTS4 NAD-dependent epimerase/dehydratase n=1 Tax=Cht... 65 2e-09
UniRef100_A4TUX1 NAD-dependent epimerase/dehydratase n=1 Tax=Mag... 65 3e-09
UniRef100_A4KUB4 TlmG n=1 Tax=Streptoalloteichus hindustanus Rep... 64 4e-09
UniRef100_B9XEZ3 NAD-dependent epimerase/dehydratase n=1 Tax=bac... 64 7e-09
UniRef100_Q1VUQ5 Sugar epimerase BlmG n=1 Tax=Psychroflexus torq... 63 9e-09
UniRef100_B8GDS4 NAD-dependent epimerase/dehydratase n=1 Tax=Met... 63 9e-09
UniRef100_A8P5B4 Putative uncharacterized protein n=1 Tax=Coprin... 62 2e-08
UniRef100_UPI000187E739 hypothetical protein MPER_11760 n=1 Tax=... 61 3e-08
UniRef100_Q2BI22 DTDP-glucose dehydratase n=1 Tax=Neptuniibacter... 61 4e-08
UniRef100_Q18EM2 Nucleoside-diphosphate-sugar epimerase (Probabl... 59 2e-07
UniRef100_B7KF75 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 55 2e-06
UniRef100_C6A080 UDP-or dTTP-glucose 4-epimerase or 4-6-dehydrat... 55 2e-06
UniRef100_B4B7N0 NAD-dependent epimerase/dehydratase n=1 Tax=Cya... 54 4e-06
UniRef100_C0UY85 Nucleoside-diphosphate-sugar epimerase n=1 Tax=... 54 5e-06
UniRef100_Q7NET6 Glr3792 protein n=1 Tax=Gloeobacter violaceus R... 54 7e-06
UniRef100_A5GEL7 NAD-dependent epimerase/dehydratase n=1 Tax=Geo... 54 7e-06
UniRef100_UPI00016C5039 NAD-dependent epimerase/dehydratase n=1 ... 53 9e-06
UniRef100_Q9K6M0 Nucleotide sugar epimerase (Biosynthesis of lip... 53 9e-06
UniRef100_Q2LVX3 GDP-L-fucose synthase n=1 Tax=Syntrophus acidit... 53 9e-06