[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV419001 MWM163g04_r
(398 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
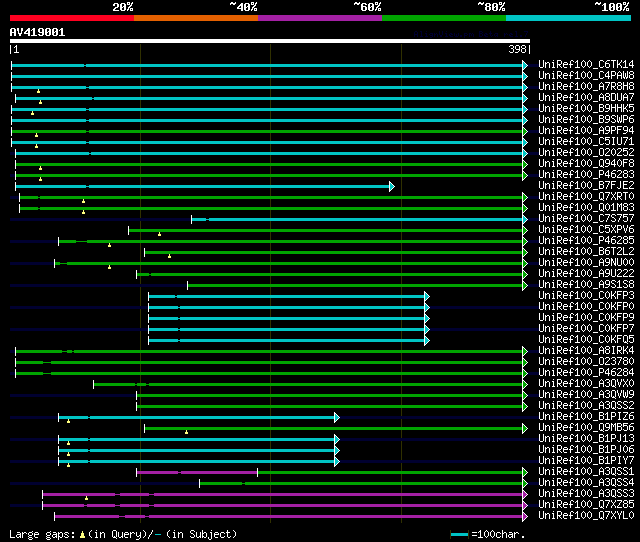
significant alignments:[graphical|details]
UniRef100_C6TK14 Putative uncharacterized protein n=1 Tax=Glycin... 231 2e-59
UniRef100_C4PAW8 Sedoheptulose-1,7-bisphosphatase n=1 Tax=Cucumi... 195 1e-48
UniRef100_A7R8H8 Chromosome undetermined scaffold_2569, whole ge... 194 2e-48
UniRef100_A8DUA7 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 186 5e-46
UniRef100_B9HHK5 Predicted protein n=1 Tax=Populus trichocarpa R... 185 2e-45
UniRef100_B9SWP6 Sedoheptulose-1,7-bisphosphatase, chloroplast, ... 183 6e-45
UniRef100_A9PF94 Predicted protein n=1 Tax=Populus trichocarpa R... 181 2e-44
UniRef100_C5IU71 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 180 4e-44
UniRef100_O20252 Sedoheptulose-1,7-bisphosphatase, chloroplastic... 176 9e-43
UniRef100_Q940F8 Sedoheptulose-bisphosphatase n=1 Tax=Arabidopsi... 174 2e-42
UniRef100_P46283 Sedoheptulose-1,7-bisphosphatase, chloroplastic... 174 2e-42
UniRef100_B7FJE2 Putative uncharacterized protein n=1 Tax=Medica... 147 3e-34
UniRef100_Q7XRT0 Os04g0234600 protein n=1 Tax=Oryza sativa Japon... 143 7e-33
UniRef100_Q01M83 H0622G10.1 protein n=3 Tax=Oryza sativa RepID=Q... 143 7e-33
UniRef100_C7S757 Fructose-1,6-bisphosphatase (Fragment) n=1 Tax=... 142 9e-33
UniRef100_C5XPV6 Putative uncharacterized protein Sb03g040100 n=... 135 2e-30
UniRef100_P46285 Sedoheptulose-1,7-bisphosphatase, chloroplastic... 135 2e-30
UniRef100_B6T2L2 Sedoheptulose-1,7-bisphosphatase n=1 Tax=Zea ma... 131 2e-29
UniRef100_A9NU00 Putative uncharacterized protein n=1 Tax=Picea ... 130 6e-29
UniRef100_A9U222 Predicted protein n=1 Tax=Physcomitrella patens... 118 2e-25
UniRef100_A9S1S8 Predicted protein n=1 Tax=Physcomitrella patens... 114 3e-24
UniRef100_C0KFP3 COSII_At3g55800 (Fragment) n=4 Tax=Lycopersicon... 109 1e-22
UniRef100_C0KFP0 COSII_At3g55800 (Fragment) n=11 Tax=Solanoideae... 108 1e-22
UniRef100_C0KFP9 COSII_At3g55800 (Fragment) n=4 Tax=Solanum RepI... 108 2e-22
UniRef100_C0KFP7 COSII_At3g55800 (Fragment) n=1 Tax=Solanum huay... 108 2e-22
UniRef100_C0KFQ5 COSII_At3g55800 (Fragment) n=1 Tax=Solanum jugl... 107 4e-22
UniRef100_A8IRK4 Sedoheptulose-1,7-bisphosphatase n=1 Tax=Chlamy... 104 3e-21
UniRef100_O23780 Sedoheptulose-1,7-biphosphatase n=1 Tax=Chlamyd... 103 8e-21
UniRef100_P46284 Sedoheptulose-1,7-bisphosphatase, chloroplastic... 103 8e-21
UniRef100_A3QVX0 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 100 4e-20
UniRef100_A3QVW9 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 100 5e-20
UniRef100_A3QSS2 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 100 5e-20
UniRef100_B1PIZ6 Chloroplast sedoheptulose-1,7-bisphosphatase (F... 99 2e-19
UniRef100_Q9MB56 Sedoheptulose-1,7-bisphosphatase n=1 Tax=Chlamy... 97 6e-19
UniRef100_B1PJ13 Chloroplast sedoheptulose-1,7-bisphosphatase (F... 96 2e-18
UniRef100_B1PJ06 Chloroplast sedoheptulose-1,7-bisphosphatase (F... 95 2e-18
UniRef100_B1PIY7 Chloroplast sedoheptulose-1,7-bisphosphatase (F... 94 5e-18
UniRef100_A3QSS1 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 81 4e-14
UniRef100_A3QSS4 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 64 4e-09
UniRef100_A3QSS3 Chloroplast sedoheptulose-1,7-bisphosphatase n=... 63 1e-08
UniRef100_Q7XZ85 Sedoheptulose-1,7-bisphosphatase (Fragment) n=1... 61 3e-08
UniRef100_Q7XYL0 Sedoheptulose-1,7 bisphosphatase (Fragment) n=1... 58 4e-07
UniRef100_B5A4J8 Chloroplast sedoheptulose-1,7-bisphosphatase (F... 57 5e-07