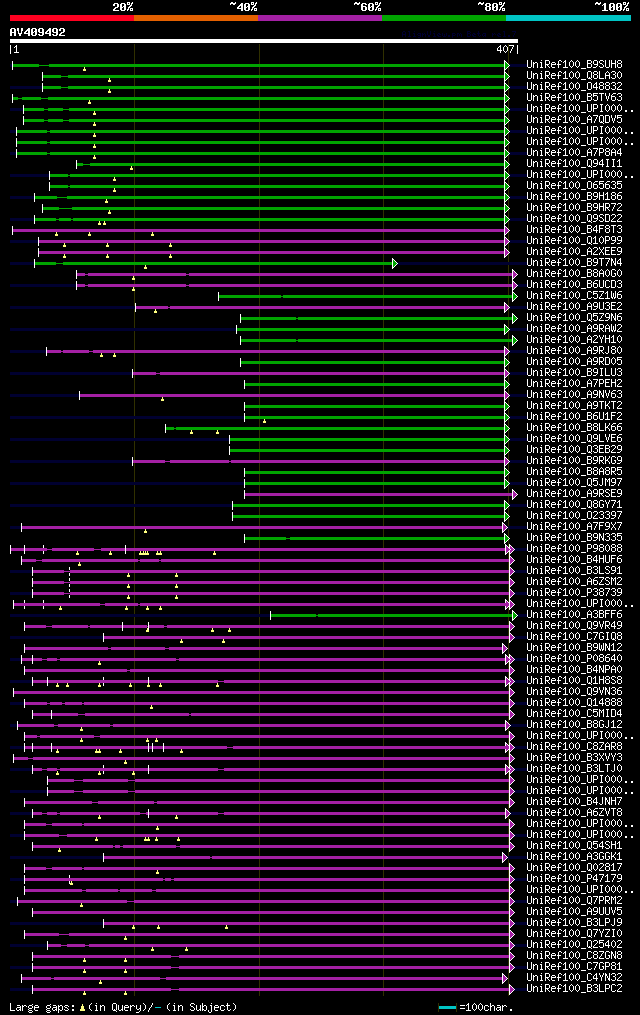
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV409492 MWL058a12_r
(407 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_B9SUH8 Putative uncharacterized protein n=1 Tax=Ricinu... 175 1e-42
UniRef100_Q8LA30 Putative senescence-associated protein 12 n=1 T... 153 7e-36
UniRef100_O48832 Putative senescence-related protein n=1 Tax=Ara... 153 7e-36
UniRef100_B5TV63 Senescence-related protein n=1 Tax=Camellia sin... 151 3e-35
UniRef100_UPI000198306C PREDICTED: hypothetical protein n=1 Tax=... 150 3e-35
UniRef100_A7QDV5 Chromosome chr4 scaffold_83, whole genome shotg... 150 3e-35
UniRef100_UPI0001982DD5 PREDICTED: hypothetical protein isoform ... 149 1e-34
UniRef100_UPI0001982DD4 PREDICTED: hypothetical protein isoform ... 149 1e-34
UniRef100_A7P8A4 Chromosome chr3 scaffold_8, whole genome shotgu... 149 1e-34
UniRef100_Q94II1 ERD7 protein (Fragment) n=1 Tax=Arabidopsis tha... 144 2e-33
UniRef100_UPI0000162B9D senescence/dehydration-associated protei... 142 2e-32
UniRef100_O65635 Putative uncharacterized protein AT4g35980 n=1 ... 142 2e-32
UniRef100_B9H186 Predicted protein n=1 Tax=Populus trichocarpa R... 134 2e-30
UniRef100_B9HR72 Predicted protein (Fragment) n=1 Tax=Populus tr... 134 4e-30
UniRef100_Q9SD22 Putative uncharacterized protein F24M12.290 n=1... 126 9e-28
UniRef100_B4F8T3 Putative uncharacterized protein n=1 Tax=Zea ma... 113 8e-24
UniRef100_Q10P99 Os03g0241900 protein n=1 Tax=Oryza sativa Japon... 107 3e-22
UniRef100_A2XEE9 Putative uncharacterized protein n=1 Tax=Oryza ... 107 3e-22
UniRef100_B9T7N4 Putative uncharacterized protein n=1 Tax=Ricinu... 88 3e-16
UniRef100_B8A0G0 Putative uncharacterized protein n=1 Tax=Zea ma... 83 1e-14
UniRef100_B6UCD3 Putative uncharacterized protein n=1 Tax=Zea ma... 83 1e-14
UniRef100_C5Z1W6 Putative uncharacterized protein Sb10g030260 n=... 81 4e-14
UniRef100_A9U3E2 Predicted protein n=1 Tax=Physcomitrella patens... 80 7e-14
UniRef100_Q5Z9N6 Os06g0717100 protein n=2 Tax=Oryza sativa RepID... 79 2e-13
UniRef100_A9RAW2 Predicted protein (Fragment) n=1 Tax=Physcomitr... 79 2e-13
UniRef100_A2YH10 Putative uncharacterized protein n=1 Tax=Oryza ... 79 2e-13
UniRef100_A9RJ80 Predicted protein n=1 Tax=Physcomitrella patens... 76 1e-12
UniRef100_A9RD05 Predicted protein (Fragment) n=1 Tax=Physcomitr... 73 9e-12
UniRef100_B9ILU3 Predicted protein (Fragment) n=1 Tax=Populus tr... 71 3e-11
UniRef100_A7PEH2 Chromosome chr11 scaffold_13, whole genome shot... 70 6e-11
UniRef100_A9NV63 Putative uncharacterized protein n=1 Tax=Picea ... 70 7e-11
UniRef100_A9TKT2 Predicted protein n=1 Tax=Physcomitrella patens... 69 1e-10
UniRef100_B6U1F2 Senescence-associated protein 12 n=1 Tax=Zea ma... 69 2e-10
UniRef100_B8LK66 Putative uncharacterized protein n=1 Tax=Picea ... 67 6e-10
UniRef100_Q9LVE6 Similarity to senescence-associated protein n=1... 65 2e-09
UniRef100_Q3EB29 Putative uncharacterized protein At3g21600.2 n=... 65 2e-09
UniRef100_B9RKG9 Putative uncharacterized protein n=1 Tax=Ricinu... 65 3e-09
UniRef100_B8A8R5 Putative uncharacterized protein n=1 Tax=Oryza ... 65 3e-09
UniRef100_Q5JM97 Os01g0723100 protein n=2 Tax=Oryza sativa Japon... 64 4e-09
UniRef100_A9RSE9 Predicted protein (Fragment) n=1 Tax=Physcomitr... 64 7e-09
UniRef100_Q8GY71 Putative uncharacterized protein At4g15450/dl37... 63 9e-09
UniRef100_O23397 Putative uncharacterized protein AT4g15450 n=1 ... 63 9e-09
UniRef100_A7F9X7 Predicted protein n=1 Tax=Sclerotinia sclerotio... 63 1e-08
UniRef100_B9N335 Predicted protein n=1 Tax=Populus trichocarpa R... 61 4e-08
UniRef100_P98088 Mucin-5AC (Fragments) n=1 Tax=Homo sapiens RepI... 60 6e-08
UniRef100_B4HUF6 GM14705 n=1 Tax=Drosophila sechellia RepID=B4HU... 60 8e-08
UniRef100_B3LS91 Cell wall integrity and stress response compone... 59 2e-07
UniRef100_A6ZSM2 Cell wall integrity and stress response compone... 59 2e-07
UniRef100_P38739 Cell wall integrity and stress response compone... 59 2e-07
UniRef100_UPI0000D9D6BA PREDICTED: mucin 5, subtype B, tracheobr... 57 5e-07
UniRef100_A3BFF6 Putative uncharacterized protein n=1 Tax=Oryza ... 57 5e-07
UniRef100_Q9VR49 Salivary gland secretion 1 n=1 Tax=Drosophila m... 57 5e-07
UniRef100_C7GIQ8 Wsc4p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 57 8e-07
UniRef100_B9WN12 Chitinase 3, putative n=1 Tax=Candida dublinien... 57 8e-07
UniRef100_P08640 Flocculation protein FLO11 n=2 Tax=Saccharomyce... 57 8e-07
UniRef100_B4NPA0 GK17577 n=1 Tax=Drosophila willistoni RepID=B4N... 56 1e-06
UniRef100_Q1H8S8 Cell surface flocculin n=1 Tax=Saccharomyces ce... 56 1e-06
UniRef100_Q9VN36 CG12586 n=1 Tax=Drosophila melanogaster RepID=Q... 56 1e-06
UniRef100_Q14888 Mucin (Fragment) n=1 Tax=Homo sapiens RepID=Q14... 56 1e-06
UniRef100_C5MID4 Predicted protein n=1 Tax=Candida tropicalis MY... 56 1e-06
UniRef100_B8GJ12 Periplasmic copper-binding n=1 Tax=Methanosphae... 56 1e-06
UniRef100_UPI0000EB4AD1 UPI0000EB4AD1 related cluster n=1 Tax=Ca... 55 2e-06
UniRef100_C8ZAR8 Muc1p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 55 2e-06
UniRef100_B3XVY3 Receptor-type protein tyrosine kinase (Fragment... 55 2e-06
UniRef100_B3LTJ0 Putative uncharacterized protein n=1 Tax=Saccha... 55 2e-06
UniRef100_UPI0001662AB7 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_UPI0001662A82 PREDICTED: hypothetical protein n=1 Tax=... 55 3e-06
UniRef100_B4JNH7 GH24138 n=1 Tax=Drosophila grimshawi RepID=B4JN... 55 3e-06
UniRef100_A6ZVT8 Cell surface flocculin n=1 Tax=Saccharomyces ce... 55 3e-06
UniRef100_UPI0000E59252 mucin 2 precursor n=1 Tax=Homo sapiens R... 54 4e-06
UniRef100_UPI0000E48EFF PREDICTED: hypothetical protein, partial... 54 4e-06
UniRef100_Q54SH1 Putative uncharacterized protein n=1 Tax=Dictyo... 54 4e-06
UniRef100_A3GGK1 Cell wall protein in family of putative glycosi... 54 4e-06
UniRef100_Q02817 Mucin-2 n=1 Tax=Homo sapiens RepID=MUC2_HUMAN 54 4e-06
UniRef100_P47179 Cell wall protein DAN4 n=1 Tax=Saccharomyces ce... 54 4e-06
UniRef100_UPI000192420E PREDICTED: hypothetical protein n=1 Tax=... 54 5e-06
UniRef100_Q7PRM2 AGAP010704-PA (Fragment) n=1 Tax=Anopheles gamb... 54 5e-06
UniRef100_A9UUV5 Predicted protein n=1 Tax=Monosiga brevicollis ... 54 5e-06
UniRef100_B3LPJ9 A-agglutinin anchorage subunit n=1 Tax=Saccharo... 54 5e-06
UniRef100_Q7YZI0 MBCTL1 (Fragment) n=1 Tax=Monosiga brevicollis ... 54 7e-06
UniRef100_Q25402 Microfilarial sheath protein SHP3 n=1 Tax=Litom... 53 9e-06
UniRef100_C8ZGN8 Wsc2p n=1 Tax=Saccharomyces cerevisiae EC1118 R... 53 9e-06
UniRef100_C7GP81 Wsc2p n=1 Tax=Saccharomyces cerevisiae JAY291 R... 53 9e-06
UniRef100_C4YN32 Putative uncharacterized protein n=1 Tax=Candid... 53 9e-06
UniRef100_B3LPC2 Cell wall integrity and stress response compone... 53 9e-06
UniRef100_A7U4Y8 Flocculin n=1 Tax=Saccharomyces cerevisiae RepI... 53 9e-06