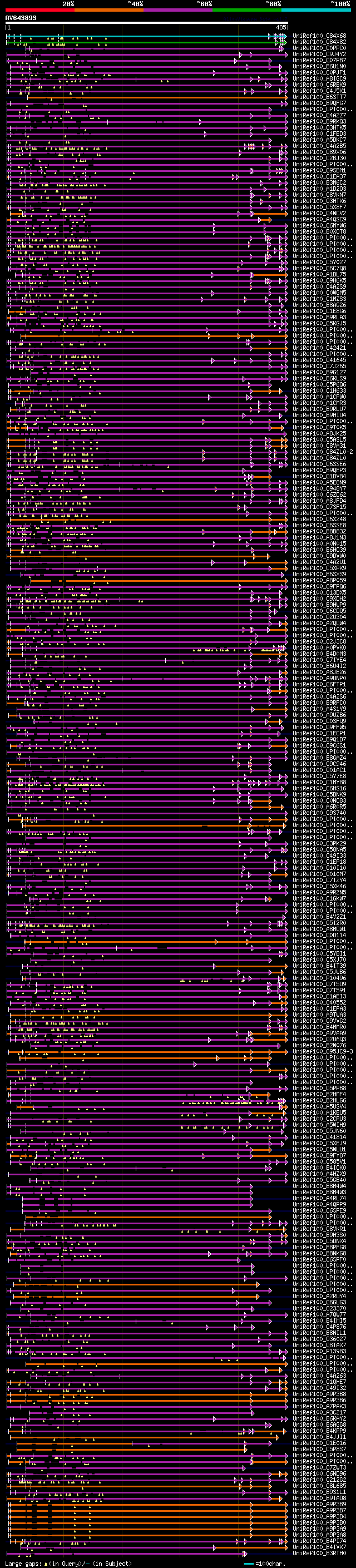
[UP]
BLASTX 2.2.22 [Sep-27-2009]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AV643893 HCL077h10_r
(485 letters)
Database: uniref100
9,526,577 sequences; 3,332,566,763 total letters
Searching..................................................done
significant alignments:[graphical|details]
UniRef100_Q84X68 Flagella membrane glycoprotein 1B n=1 Tax=Chlam... 308 1e-82
UniRef100_Q84X82 Flagella membrane glycoprotein n=1 Tax=Chlamydo... 202 9e-51
UniRef100_C0PPC0 Putative uncharacterized protein n=1 Tax=Zea ma... 82 1e-14
UniRef100_C9J4Y2 Putative uncharacterized protein ENSP0000041026... 82 2e-14
UniRef100_Q07PB7 Peptidase C14, caspase catalytic subunit p20 n=... 80 9e-14
UniRef100_B6U1N0 Putative uncharacterized protein n=1 Tax=Zea ma... 79 1e-13
UniRef100_C0PJF1 Putative uncharacterized protein n=1 Tax=Zea ma... 79 2e-13
UniRef100_A8IGC9 Fibrocystin-L-like protein n=1 Tax=Chlamydomona... 79 2e-13
UniRef100_C6RBK9 Iron-sulfur cluster-binding protein n=1 Tax=Cor... 78 3e-13
UniRef100_C4J5K1 Putative uncharacterized protein n=1 Tax=Zea ma... 78 3e-13
UniRef100_B6STT7 Extensin n=1 Tax=Zea mays RepID=B6STT7_MAIZE 78 3e-13
UniRef100_B9QFG7 Subtilisin-like protein TgSUB1, putative n=1 Ta... 78 3e-13
UniRef100_UPI000151B778 hypothetical protein PGUG_03728 n=1 Tax=... 77 5e-13
UniRef100_Q4A2Z7 Putative membrane protein n=2 Tax=Emiliania hux... 77 5e-13
UniRef100_B9RKQ3 ATP binding protein, putative n=1 Tax=Ricinus c... 77 5e-13
UniRef100_Q3HTK5 Pherophorin-C2 protein n=1 Tax=Chlamydomonas re... 77 6e-13
UniRef100_C1FED3 Predicted protein n=1 Tax=Micromonas sp. RCC299... 77 6e-13
UniRef100_A5DKC7 Putative uncharacterized protein n=1 Tax=Pichia... 77 6e-13
UniRef100_Q4A2B5 Putative membrane protein n=1 Tax=Emiliania hux... 77 8e-13
UniRef100_Q89X06 Blr0521 protein n=1 Tax=Bradyrhizobium japonicu... 77 8e-13
UniRef100_C2BJ30 Fe-S oxidoreductase n=1 Tax=Corynebacterium pse... 77 8e-13
UniRef100_UPI0000DB7674 PREDICTED: hypothetical protein n=1 Tax=... 76 1e-12
UniRef100_Q9SBM1 Hydroxyproline-rich glycoprotein DZ-HRGP n=1 Ta... 76 1e-12
UniRef100_C1EA37 Predicted protein n=1 Tax=Micromonas sp. RCC299... 76 1e-12
UniRef100_B3M6C2 GF24314 n=1 Tax=Drosophila ananassae RepID=B3M6... 76 1e-12
UniRef100_A1D2Q3 Conserved proline-rich protein n=1 Tax=Neosarto... 76 1e-12
UniRef100_Q8VKN7 Putative uncharacterized protein n=1 Tax=Mycoba... 76 1e-12
UniRef100_Q3HTK6 Pherophorin-C1 protein n=1 Tax=Chlamydomonas re... 76 1e-12
UniRef100_C5X8F7 Putative uncharacterized protein Sb02g032910 n=... 76 1e-12
UniRef100_Q4WCV2 Actin associated protein Wsp1, putative n=1 Tax... 76 1e-12
UniRef100_A4QSC9 Predicted protein n=1 Tax=Magnaporthe grisea Re... 76 1e-12
UniRef100_Q6MYW6 Basic proline-rich protein n=1 Tax=Aspergillus ... 75 2e-12
UniRef100_B0XQT8 Conserved proline-rich protein n=2 Tax=Aspergil... 75 2e-12
UniRef100_UPI0001A5EB4A PREDICTED: hypothetical protein FLJ22184... 75 2e-12
UniRef100_UPI0001925155 PREDICTED: hypothetical protein, partial... 75 2e-12
UniRef100_UPI00015B4CAB PREDICTED: hypothetical protein n=1 Tax=... 75 2e-12
UniRef100_UPI00015DF937 CDNA: FLJ22184 fis, clone HRC00983. n=1 ... 75 2e-12
UniRef100_C5Y027 Putative uncharacterized protein Sb04g010730 n=... 75 2e-12
UniRef100_Q6C7Q8 YALI0D26191p n=1 Tax=Yarrowia lipolytica RepID=... 75 2e-12
UniRef100_A1DL75 Actin associated protein Wsp1, putative n=1 Tax... 75 2e-12
UniRef100_Q9H6K5 Putative uncharacterized protein FLJ22184 n=1 T... 75 2e-12
UniRef100_Q4A2S9 Putative membrane protein n=2 Tax=Emiliania hux... 75 3e-12
UniRef100_C0WGM5 Fe-S oxidoreductase n=1 Tax=Corynebacterium acc... 75 3e-12
UniRef100_C1MZS3 Predicted protein n=1 Tax=Micromonas pusilla CC... 75 3e-12
UniRef100_B8AG26 Putative uncharacterized protein n=1 Tax=Oryza ... 75 3e-12
UniRef100_C1E8G6 Predicted protein n=1 Tax=Micromonas sp. RCC299... 74 4e-12
UniRef100_B9RLA3 ATP binding protein, putative n=1 Tax=Ricinus c... 74 4e-12
UniRef100_Q5KGJ5 Putative uncharacterized protein n=1 Tax=Filoba... 74 4e-12
UniRef100_UPI0001B55E13 putative chaplin n=1 Tax=Streptomyces sp... 74 5e-12
UniRef100_UPI000192638B PREDICTED: hypothetical protein n=1 Tax=... 74 5e-12
UniRef100_UPI0001925A99 PREDICTED: hypothetical protein, partial... 74 5e-12
UniRef100_Q42421 Chitinase n=1 Tax=Beta vulgaris subsp. vulgaris... 74 5e-12
UniRef100_UPI0000E1257F Os05g0516400 n=1 Tax=Oryza sativa Japoni... 74 7e-12
UniRef100_Q41645 Extensin (Fragment) n=1 Tax=Volvox carteri RepI... 74 7e-12
UniRef100_C7J265 Os05g0516400 protein (Fragment) n=1 Tax=Oryza s... 74 7e-12
UniRef100_B9G127 Putative uncharacterized protein n=1 Tax=Oryza ... 74 7e-12
UniRef100_B6KLS9 Voltage gated chloride channel domain-containin... 74 7e-12
UniRef100_C5P6Q6 WH1 domain containing protein n=1 Tax=Coccidioi... 74 7e-12
UniRef100_C1H633 Predicted protein n=1 Tax=Paracoccidioides bras... 74 7e-12
UniRef100_A1CPW0 Conserved proline-rich protein n=1 Tax=Aspergil... 74 7e-12
UniRef100_A1CMR3 Actin associated protein Wsp1, putative n=1 Tax... 74 7e-12
UniRef100_B9RLU7 Putative uncharacterized protein n=1 Tax=Ricinu... 73 9e-12
UniRef100_B9HIU4 Predicted protein (Fragment) n=1 Tax=Populus tr... 73 9e-12
UniRef100_UPI0000E12BCF Os07g0596300 n=1 Tax=Oryza sativa Japoni... 73 1e-11
UniRef100_Q9T0K5 Extensin-like protein n=1 Tax=Arabidopsis thali... 73 1e-11
UniRef100_A8JK25 Flagella membrane glycoprotein (Fragment) n=1 T... 73 1e-11
UniRef100_Q5ASL5 Putative uncharacterized protein n=1 Tax=Emeric... 73 1e-11
UniRef100_C8VA31 Putative uncharacterized protein n=1 Tax=Asperg... 73 1e-11
UniRef100_Q84ZL0-2 Isoform 2 of Formin-like protein 5 n=1 Tax=Or... 73 1e-11
UniRef100_Q84ZL0 Formin-like protein 5 n=1 Tax=Oryza sativa Japo... 73 1e-11
UniRef100_Q6SSE6 Plus agglutinin n=1 Tax=Chlamydomonas reinhardt... 72 1e-11
UniRef100_B9QEP3 Chloride channel protein k, putative n=1 Tax=To... 72 1e-11
UniRef100_Q1DV84 Putative uncharacterized protein n=1 Tax=Coccid... 72 1e-11
UniRef100_A5E8N9 Putative uncharacterized protein n=1 Tax=Bradyr... 72 2e-11
UniRef100_Q948Y7 VMP3 protein n=1 Tax=Volvox carteri f. nagarien... 72 2e-11
UniRef100_Q6ZD62 Os08g0108300 protein n=1 Tax=Oryza sativa Japon... 72 2e-11
UniRef100_A8JFD4 Glyoxal or galactose oxidase (Fragment) n=1 Tax... 72 2e-11
UniRef100_Q7SF15 WASP-like pretein las17p n=1 Tax=Neurospora cra... 72 2e-11
UniRef100_UPI0001A7B3C7 unknown protein n=1 Tax=Arabidopsis thal... 72 2e-11
UniRef100_Q6X248 UL36 very large tegument protein n=1 Tax=Bovine... 72 2e-11
UniRef100_Q6SSE8 Minus agglutinin n=1 Tax=Chlamydomonas reinhard... 72 2e-11
UniRef100_B8B832 Putative uncharacterized protein n=1 Tax=Oryza ... 72 2e-11
UniRef100_A8J1N3 Cell wall protein pherophorin-C10 (Fragment) n=... 72 2e-11
UniRef100_A0N015 Vegetative cell wall protein n=1 Tax=Chlamydomo... 72 2e-11
UniRef100_B6HQ39 Pc22g23920 protein n=1 Tax=Penicillium chrysoge... 72 2e-11
UniRef100_Q9DVW0 PxORF73 peptide n=1 Tax=Plutella xylostella gra... 71 3e-11
UniRef100_Q4A2U1 Putative membrane protein n=2 Tax=Emiliania hux... 71 3e-11
UniRef100_C5XPK9 Putative uncharacterized protein Sb03g026730 n=... 71 3e-11
UniRef100_B6SXS9 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 71 3e-11
UniRef100_A8P059 Pherophorin-dz1 protein, putative n=1 Tax=Brugi... 71 3e-11
UniRef100_Q9FPQ6 Vegetative cell wall protein gp1 n=1 Tax=Chlamy... 71 3e-11
UniRef100_Q13DX5 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Bi... 71 4e-11
UniRef100_Q9XDH2 Proline-rich mucin homolog n=1 Tax=Mycobacteriu... 71 4e-11
UniRef100_B9HWP9 Predicted protein (Fragment) n=1 Tax=Populus tr... 71 4e-11
UniRef100_Q6CDQ5 YALI0B22110p n=1 Tax=Yarrowia lipolytica RepID=... 71 4e-11
UniRef100_Q2U304 Predicted protein n=1 Tax=Aspergillus oryzae Re... 71 4e-11
UniRef100_A2QQW4 Contig An08c0110, complete genome n=1 Tax=Asper... 71 4e-11
UniRef100_UPI0001982D5B PREDICTED: hypothetical protein n=1 Tax=... 70 6e-11
UniRef100_UPI0000D9B0F8 PREDICTED: similar to mucin 7, salivary ... 70 6e-11
UniRef100_Q2J3C8 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Ha... 70 6e-11
UniRef100_A0PVK0 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 70 6e-11
UniRef100_B4D0M3 Putative uncharacterized protein n=1 Tax=Chthon... 70 6e-11
UniRef100_C7IYE4 Os02g0557000 protein (Fragment) n=1 Tax=Oryza s... 70 6e-11
UniRef100_B6U4I2 Putative uncharacterized protein n=1 Tax=Zea ma... 70 6e-11
UniRef100_A8JE26 Predicted protein (Fragment) n=1 Tax=Chlamydomo... 70 6e-11
UniRef100_A9UNP0 SH3, pleckstrin-like and PDZ/DHR/GLGF domain-co... 70 6e-11
UniRef100_Q6FTP1 Similar to uniprot|P37370 Saccharomyces cerevis... 70 6e-11
UniRef100_UPI00019829A2 PREDICTED: hypothetical protein n=1 Tax=... 70 7e-11
UniRef100_Q4A2S6 Putative membrane protein n=2 Tax=Emiliania hux... 70 7e-11
UniRef100_B9RPC0 LRX1, putative n=1 Tax=Ricinus communis RepID=B... 70 7e-11
UniRef100_A4S1Y9 Predicted protein n=1 Tax=Ostreococcus lucimari... 70 7e-11
UniRef100_A9UZB6 Predicted protein n=1 Tax=Monosiga brevicollis ... 70 7e-11
UniRef100_C0SFQ9 Putative uncharacterized protein n=1 Tax=Paraco... 70 7e-11
UniRef100_Q9FFW5 Similarity to protein kinase n=1 Tax=Arabidopsi... 70 9e-11
UniRef100_C1ECP1 Resistance-nodulation-cell division superfamily... 70 9e-11
UniRef100_B9Q1D7 Chloride channel, putative n=1 Tax=Toxoplasma g... 70 9e-11
UniRef100_Q9C6S1 Formin-like protein 14 n=1 Tax=Arabidopsis thal... 70 9e-11
UniRef100_UPI00017390C7 AGP19 (ARABINOGALACTAN-PROTEIN 19) n=1 T... 69 1e-10
UniRef100_B8GAZ4 Putative uncharacterized protein n=1 Tax=Chloro... 69 1e-10
UniRef100_Q9C946 Putative uncharacterized protein T7P1.21 n=1 Ta... 69 1e-10
UniRef100_Q01AC1 Meltrins, fertilins and related Zn-dependent me... 69 1e-10
UniRef100_C5Y7E8 Putative uncharacterized protein Sb05g025945 (F... 69 1e-10
UniRef100_C1MY88 Predicted protein n=1 Tax=Micromonas pusilla CC... 69 1e-10
UniRef100_C6HS16 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 69 1e-10
UniRef100_C5DNK9 KLTH0G17886p n=1 Tax=Lachancea thermotolerans C... 69 1e-10
UniRef100_C0NQ83 Putative uncharacterized protein n=1 Tax=Ajello... 69 1e-10
UniRef100_A6R0R5 Predicted protein n=1 Tax=Ajellomyces capsulatu... 69 1e-10
UniRef100_Q9S740 Lysine-rich arabinogalactan protein 19 n=1 Tax=... 69 1e-10
UniRef100_UPI00015B501E PREDICTED: hypothetical protein n=1 Tax=... 69 2e-10
UniRef100_UPI0000E4981C PREDICTED: hypothetical protein n=1 Tax=... 69 2e-10
UniRef100_UPI0000E24F27 PREDICTED: hypothetical protein n=1 Tax=... 69 2e-10
UniRef100_UPI0000E11F6B Os03g0190000 n=1 Tax=Oryza sativa Japoni... 69 2e-10
UniRef100_C3PK29 Putative Fe-S oxidoreductase n=1 Tax=Corynebact... 69 2e-10
UniRef100_Q58NA5 Plus agglutinin (Fragment) n=1 Tax=Chlamydomona... 69 2e-10
UniRef100_Q49I33 120 kDa pistil extensin-like protein (Fragment)... 69 2e-10
UniRef100_Q1EP18 Protein kinase family protein n=1 Tax=Musa balb... 69 2e-10
UniRef100_Q10I10 Os03g0568800 protein n=1 Tax=Oryza sativa Japon... 69 2e-10
UniRef100_Q010M7 Predicted membrane protein (Patched superfamily... 69 2e-10
UniRef100_C7IZY4 Os03g0189900 protein n=1 Tax=Oryza sativa Japon... 69 2e-10
UniRef100_C5XK46 Putative uncharacterized protein Sb03g034710 n=... 69 2e-10
UniRef100_A9RZN5 Predicted protein n=1 Tax=Physcomitrella patens... 69 2e-10
UniRef100_C1GKW7 Predicted protein n=1 Tax=Paracoccidioides bras... 69 2e-10
UniRef100_UPI00006A1DCC UPI00006A1DCC related cluster n=1 Tax=Xe... 69 2e-10
UniRef100_UPI00006A1DCA UPI00006A1DCA related cluster n=1 Tax=Xe... 69 2e-10
UniRef100_B4V2Z1 Serine/threonine protein kinase n=1 Tax=Strepto... 69 2e-10
UniRef100_Q5I2R0 Minus agglutinin n=1 Tax=Chlamydomonas incerta ... 69 2e-10
UniRef100_A8MQW1 Uncharacterized protein At1g44191.1 n=1 Tax=Ara... 69 2e-10
UniRef100_Q0D114 Predicted protein n=1 Tax=Aspergillus terreus N... 69 2e-10
UniRef100_UPI0001926FBD PREDICTED: hypothetical protein n=1 Tax=... 68 3e-10
UniRef100_UPI000069F8DC UPI000069F8DC related cluster n=1 Tax=Xe... 68 3e-10
UniRef100_C5YBI1 Putative uncharacterized protein Sb06g021550 n=... 68 3e-10
UniRef100_C5XJ70 Putative uncharacterized protein Sb03g034040 n=... 68 3e-10
UniRef100_B4IT39 GE18257 n=1 Tax=Drosophila yakuba RepID=B4IT39_... 68 3e-10
UniRef100_C5JWB6 Putative uncharacterized protein n=1 Tax=Ajello... 68 3e-10
UniRef100_P10496 Glycine-rich cell wall structural protein 1.8 n... 68 3e-10
UniRef100_Q7T5D9 Very large tegument protein n=1 Tax=Macacine he... 68 4e-10
UniRef100_Q7T591 Large tegument protein n=1 Tax=Macacine herpesv... 68 4e-10
UniRef100_C1AEI3 Putative uncharacterized protein n=1 Tax=Gemmat... 68 4e-10
UniRef100_Q40552 Pistil extensin like protein (Fragment) n=1 Tax... 68 4e-10
UniRef100_Q1EPA3 Protein kinase family protein n=1 Tax=Musa acum... 68 4e-10
UniRef100_A9TWA3 Predicted protein n=1 Tax=Physcomitrella patens... 68 4e-10
UniRef100_Q9VVG2 CG13731 n=1 Tax=Drosophila melanogaster RepID=Q... 68 4e-10
UniRef100_B4MMR0 GK17563 n=1 Tax=Drosophila willistoni RepID=B4M... 68 4e-10
UniRef100_A9VAA9 Predicted protein n=1 Tax=Monosiga brevicollis ... 68 4e-10
UniRef100_Q2U6Q3 Actin regulatory protein n=1 Tax=Aspergillus or... 68 4e-10
UniRef100_B2W076 Predicted protein n=1 Tax=Pyrenophora tritici-r... 68 4e-10
UniRef100_Q95JC9-3 Isoform 3 of Basic proline-rich protein n=1 T... 68 4e-10
UniRef100_UPI000186983E hypothetical protein BRAFLDRAFT_104497 n... 67 5e-10
UniRef100_UPI0001868DB9 hypothetical protein BRAFLDRAFT_129698 n... 67 5e-10
UniRef100_UPI00015B541C PREDICTED: hypothetical protein n=1 Tax=... 67 5e-10
UniRef100_UPI0000D9EA10 PREDICTED: hypothetical protein n=1 Tax=... 67 5e-10
UniRef100_UPI000023F096 hypothetical protein FG08656.1 n=1 Tax=G... 67 5e-10
UniRef100_Q5PPB8 UL36 tegument protein n=2 Tax=Suid herpesvirus ... 67 5e-10
UniRef100_B2HMF4 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 67 5e-10
UniRef100_B2HLG6 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 67 5e-10
UniRef100_A5USV4 Putative uncharacterized protein n=1 Tax=Roseif... 67 5e-10
UniRef100_A1KEU5 PE-PGRS family protein n=3 Tax=Mycobacterium bo... 67 5e-10
UniRef100_C2CRU3 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 67 5e-10
UniRef100_A5WIH9 PE-PGRS family protein n=2 Tax=Mycobacterium tu... 67 5e-10
UniRef100_Q5JN60 Os01g0750600 protein n=1 Tax=Oryza sativa Japon... 67 5e-10
UniRef100_Q41814 Hydroxyproline-rich glycoprotein n=1 Tax=Zea ma... 67 5e-10
UniRef100_C5XEJ9 Putative uncharacterized protein Sb03g029150 n=... 67 5e-10
UniRef100_C5WUU1 Putative uncharacterized protein Sb01g044570 n=... 67 5e-10
UniRef100_B9FY87 Putative uncharacterized protein n=1 Tax=Oryza ... 67 5e-10
UniRef100_Q585V1 Putative uncharacterized protein n=1 Tax=Trypan... 67 5e-10
UniRef100_B4IQK0 GM19288 n=1 Tax=Drosophila sechellia RepID=B4IQ... 67 5e-10
UniRef100_A4HZX9 Putative uncharacterized protein n=1 Tax=Leishm... 67 5e-10
UniRef100_C5GB40 Actin associated protein Wsp1 n=1 Tax=Ajellomyc... 67 5e-10
UniRef100_B8M4W4 Actin associated protein Wsp1, putative n=1 Tax... 67 5e-10
UniRef100_B8M4W3 Actin associated protein Wsp1, putative n=1 Tax... 67 5e-10
UniRef100_A4RL74 Putative uncharacterized protein n=1 Tax=Magnap... 67 5e-10
UniRef100_A4QPP9 Putative uncharacterized protein n=1 Tax=Magnap... 67 5e-10
UniRef100_Q6SPE9 Atherin n=1 Tax=Oryctolagus cuniculus RepID=SAM... 67 5e-10
UniRef100_UPI00019841A9 PREDICTED: hypothetical protein n=1 Tax=... 67 6e-10
UniRef100_UPI0001983015 PREDICTED: hypothetical protein n=1 Tax=... 67 6e-10
UniRef100_Q8VKR1 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 67 6e-10
UniRef100_B9H3S0 Predicted protein (Fragment) n=1 Tax=Populus tr... 67 6e-10
UniRef100_C5DNX4 ZYRO0A12386p n=1 Tax=Zygosaccharomyces rouxii C... 67 6e-10
UniRef100_B8PFG8 Predicted protein n=1 Tax=Postia placenta Mad-6... 67 6e-10
UniRef100_B8NKG8 Actin associated protein Wsp1, putative n=1 Tax... 67 6e-10
UniRef100_Q6SPF0 Atherin n=1 Tax=Homo sapiens RepID=SAMD1_HUMAN 67 6e-10
UniRef100_UPI0001A7B3CA lipid binding / structural constituent o... 67 8e-10
UniRef100_UPI0001A7B0BB lipid binding / structural constituent o... 67 8e-10
UniRef100_UPI0001982E0B PREDICTED: hypothetical protein n=1 Tax=... 67 8e-10
UniRef100_UPI0001A2BBDD hypothetical protein LOC100009637 n=1 Ta... 67 8e-10
UniRef100_UPI000013E9DD mucin 7, secreted precursor n=1 Tax=Homo... 67 8e-10
UniRef100_A2RUY4 Zgc:158395 protein n=1 Tax=Danio rerio RepID=A2... 67 8e-10
UniRef100_Q6GUG3 Serine/proline-rich repeat protein n=2 Tax=Lupi... 67 8e-10
UniRef100_O23370 Cell wall protein like n=1 Tax=Arabidopsis thal... 67 8e-10
UniRef100_A7QW77 Chromosome chr3 scaffold_199, whole genome shot... 67 8e-10
UniRef100_B4IMI5 GM16182 n=1 Tax=Drosophila sechellia RepID=B4IM... 67 8e-10
UniRef100_Q4P876 Putative uncharacterized protein n=1 Tax=Ustila... 67 8e-10
UniRef100_B8NIL1 Proline-rich, actin-associated protein Vrp1, pu... 67 8e-10
UniRef100_O36027 Wiskott-Aldrich syndrome protein homolog 1 n=1 ... 67 8e-10
UniRef100_Q8TAX7 Mucin-7 n=1 Tax=Homo sapiens RepID=MUC7_HUMAN 67 8e-10
UniRef100_P13983 Extensin n=1 Tax=Nicotiana tabacum RepID=EXTN_T... 67 8e-10
UniRef100_UPI0001B46962 PE-PGRS family protein n=1 Tax=Mycobacte... 66 1e-09
UniRef100_UPI0001982977 PREDICTED: hypothetical protein n=1 Tax=... 66 1e-09
UniRef100_UPI00016E92A6 UPI00016E92A6 related cluster n=1 Tax=Ta... 66 1e-09
UniRef100_Q4A263 Putative membrane protein n=1 Tax=Emiliania hux... 66 1e-09
UniRef100_Q1QHE7 OmpA/MotB n=1 Tax=Nitrobacter hamburgensis X14 ... 66 1e-09
UniRef100_Q49I32 120 kDa pistil extensin-like protein (Fragment)... 66 1e-09
UniRef100_A9P3B8 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3B6 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A7PAK3 Chromosome chr14 scaffold_9, whole genome shotg... 66 1e-09
UniRef100_A3C217 Putative uncharacterized protein n=1 Tax=Oryza ... 66 1e-09
UniRef100_B6KAY2 Putative uncharacterized protein n=2 Tax=Toxopl... 66 1e-09
UniRef100_B6AGG8 Putative uncharacterized protein n=1 Tax=Crypto... 66 1e-09
UniRef100_B4KRP9 GI19648 n=1 Tax=Drosophila mojavensis RepID=B4K... 66 1e-09
UniRef100_B4JJI1 GH12253 n=1 Tax=Drosophila grimshawi RepID=B4JJ... 66 1e-09
UniRef100_Q1E016 Predicted protein n=1 Tax=Coccidioides immitis ... 66 1e-09
UniRef100_C5P8S7 Pherophorin-dz1 protein, putative n=2 Tax=Cocci... 66 1e-09
UniRef100_UPI0001982EE5 PREDICTED: hypothetical protein n=1 Tax=... 66 1e-09
UniRef100_UPI0001868875 hypothetical protein BRAFLDRAFT_103538 n... 66 1e-09
UniRef100_Q7ZWT3 Sf1 protein n=1 Tax=Xenopus laevis RepID=Q7ZWT3... 66 1e-09
UniRef100_Q6ND96 Possible OmpA family member n=1 Tax=Rhodopseudo... 66 1e-09
UniRef100_Q212G2 Peptidase C14, caspase catalytic subunit p20 n=... 66 1e-09
UniRef100_Q8L685 Pherophorin-dz1 protein n=1 Tax=Volvox carteri ... 66 1e-09
UniRef100_B9S1L1 DNA binding protein, putative n=1 Tax=Ricinus c... 66 1e-09
UniRef100_B9IAD8 Predicted protein n=1 Tax=Populus trichocarpa R... 66 1e-09
UniRef100_A9P3B9 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3B7 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3B4 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3B0 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_A9P3A9 Anther-specific proline rich protein n=4 Tax=Br... 66 1e-09
UniRef100_A9P3A8 Anther-specific proline rich protein n=1 Tax=Br... 66 1e-09
UniRef100_B4PI74 GE21449 n=1 Tax=Drosophila yakuba RepID=B4PI74_... 66 1e-09
UniRef100_B4IVK7 GE14960 n=1 Tax=Drosophila yakuba RepID=B4IVK7_... 66 1e-09
UniRef100_B3RTH0 Putative uncharacterized protein n=1 Tax=Tricho... 66 1e-09
UniRef100_B3NC39 GG15230 n=1 Tax=Drosophila erecta RepID=B3NC39_... 66 1e-09
UniRef100_Q6AHW2 Protease-1 (PRT1),, putative (Fragment) n=1 Tax... 66 1e-09
UniRef100_Q6AHS6 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 66 1e-09
UniRef100_C5FD36 Proline-rich protein LAS17 n=1 Tax=Microsporum ... 66 1e-09
UniRef100_A1DF14 Cell wall protein, putative n=1 Tax=Neosartorya... 66 1e-09
UniRef100_Q6ZQP7 Uncharacterized protein LOC284861 n=1 Tax=Homo ... 66 1e-09
UniRef100_Q2GVT8 Protein transport protein SEC31 n=1 Tax=Chaetom... 66 1e-09
UniRef100_UPI0001901195 hypothetical protein Mtub0_02808 n=1 Tax... 65 2e-09
UniRef100_UPI00006A0472 UPI00006A0472 related cluster n=1 Tax=Xe... 65 2e-09
UniRef100_UPI00016E98C2 UPI00016E98C2 related cluster n=1 Tax=Ta... 65 2e-09
UniRef100_Q7U160 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 65 2e-09
UniRef100_Q79FV7 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium tu... 65 2e-09
UniRef100_B2HSS6 PE-PGRS family protein, PE_PGRS50 n=1 Tax=Mycob... 65 2e-09
UniRef100_B2HFX6 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 65 2e-09
UniRef100_A8LE29 Putative uncharacterized protein n=1 Tax=Franki... 65 2e-09
UniRef100_A1KGW5 PE-PGRS family protein n=2 Tax=Mycobacterium bo... 65 2e-09
UniRef100_Q7D974 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 65 2e-09
UniRef100_C6DVD8 PE-PGRS family protein n=2 Tax=Mycobacterium tu... 65 2e-09
UniRef100_C4RC07 Glutamyl-tRNA reductase (Fragment) n=1 Tax=Micr... 65 2e-09
UniRef100_C0VX39 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 65 2e-09
UniRef100_B5JHK2 Translation initiation factor IF-2 n=1 Tax=Verr... 65 2e-09
UniRef100_A5WKK2 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 65 2e-09
UniRef100_Q9SL09 Putative uncharacterized protein At2g05580 n=1 ... 65 2e-09
UniRef100_Q9LT74 Similarity to late embryogenesis abundant prote... 65 2e-09
UniRef100_Q9ASK4 Os01g0110500 protein n=1 Tax=Oryza sativa Japon... 65 2e-09
UniRef100_Q7Y215 Putative uncharacterized protein At2g05580 n=1 ... 65 2e-09
UniRef100_Q7XUL1 Os04g0498700 protein n=1 Tax=Oryza sativa Japon... 65 2e-09
UniRef100_Q4KXE0 Cold acclimation induced protein 2-1 n=1 Tax=Tr... 65 2e-09
UniRef100_Q43558 Proline rich protein n=1 Tax=Medicago sativa Re... 65 2e-09
UniRef100_C5XS10 Putative uncharacterized protein Sb04g033190 n=... 65 2e-09
UniRef100_C4IYP5 Putative uncharacterized protein n=1 Tax=Zea ma... 65 2e-09
UniRef100_C1EF12 Predicted protein n=1 Tax=Micromonas sp. RCC299... 65 2e-09
UniRef100_B8B912 Putative uncharacterized protein n=1 Tax=Oryza ... 65 2e-09
UniRef100_B8ACV5 Putative uncharacterized protein n=1 Tax=Oryza ... 65 2e-09
UniRef100_B6TRM4 Putative uncharacterized protein n=1 Tax=Zea ma... 65 2e-09
UniRef100_Q95UQ2 Subtilisin-like protein n=1 Tax=Toxoplasma gond... 65 2e-09
UniRef100_A3LVW7 Predicted protein n=1 Tax=Pichia stipitis RepID... 65 2e-09
UniRef100_Q6ZQP7-2 Isoform 2 of Uncharacterized protein LOC28486... 65 2e-09
UniRef100_Q95JC9 Parotid hormone n=1 Tax=Sus scrofa RepID=PRP_PIG 65 2e-09
UniRef100_UPI00018664BA hypothetical protein BRAFLDRAFT_91133 n=... 65 2e-09
UniRef100_UPI0000E59252 mucin 2 precursor n=1 Tax=Homo sapiens R... 65 2e-09
UniRef100_UPI0000E49610 PREDICTED: hypothetical protein, partial... 65 2e-09
UniRef100_UPI00016E1896 UPI00016E1896 related cluster n=1 Tax=Ta... 65 2e-09
UniRef100_B2HMJ7 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 65 2e-09
UniRef100_A3Q834 Putative uncharacterized protein n=2 Tax=Mycoba... 65 2e-09
UniRef100_A1UNN7 Putative uncharacterized protein n=1 Tax=Mycoba... 65 2e-09
UniRef100_Q41719 Hydroxyproline-rich glycoprotein n=1 Tax=Zea di... 65 2e-09
UniRef100_B8A9G0 Putative uncharacterized protein n=1 Tax=Oryza ... 65 2e-09
UniRef100_A4S6G3 Predicted protein (Fragment) n=1 Tax=Ostreococc... 65 2e-09
UniRef100_A3KD22 Leucine-rich repeat/extensin 1 n=1 Tax=Nicotian... 65 2e-09
UniRef100_Q4QE97 Formin A n=1 Tax=Leishmania major RepID=Q4QE97_... 65 2e-09
UniRef100_B3MVK1 GF23569 n=1 Tax=Drosophila ananassae RepID=B3MV... 65 2e-09
UniRef100_C8VTC6 Proline-rich, actin-associated protein Vrp1, pu... 65 2e-09
UniRef100_Q18DZ3 Probable cell surface glycoprotein n=1 Tax=Halo... 65 2e-09
UniRef100_C4KK37 Putative uncharacterized protein n=1 Tax=Sulfol... 65 2e-09
UniRef100_Q02817 Mucin-2 n=1 Tax=Homo sapiens RepID=MUC2_HUMAN 65 2e-09
UniRef100_UPI0000DD9486 Os08g0280200 n=1 Tax=Oryza sativa Japoni... 65 3e-09
UniRef100_UPI0000546264 splicing factor 1 n=1 Tax=Danio rerio Re... 65 3e-09
UniRef100_UPI00016E7DEA UPI00016E7DEA related cluster n=1 Tax=Ta... 65 3e-09
UniRef100_B3DKQ7 Sf1 protein n=1 Tax=Danio rerio RepID=B3DKQ7_DANRE 65 3e-09
UniRef100_Q0GYD2 Putative uncharacterized protein n=1 Tax=Plutel... 65 3e-09
UniRef100_Q0RSN4 Putative uncharacterized protein n=1 Tax=Franki... 65 3e-09
UniRef100_B2HT48 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 65 3e-09
UniRef100_A1UJA1 PE-PGRS family protein n=2 Tax=Mycobacterium Re... 65 3e-09
UniRef100_A0PM18 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 65 3e-09
UniRef100_Q49I31 120 kDa pistil extensin-like protein (Fragment)... 65 3e-09
UniRef100_Q49I29 120 kDa pistil extensin-like protein (Fragment)... 65 3e-09
UniRef100_Q49I27 120 kDa pistil extensin-like protein (Fragment)... 65 3e-09
UniRef100_Q42366 Hydroxyproline-rich Glycoprotein (HRGP) n=1 Tax... 65 3e-09
UniRef100_B4L805 GI11013 n=1 Tax=Drosophila mojavensis RepID=B4L... 65 3e-09
UniRef100_A0EFA7 Chromosome undetermined scaffold_93, whole geno... 65 3e-09
UniRef100_Q6CW84 KLLA0B06039p n=1 Tax=Kluyveromyces lactis RepID... 65 3e-09
UniRef100_C7YMP5 Putative uncharacterized protein n=1 Tax=Nectri... 65 3e-09
UniRef100_C4YB97 Putative uncharacterized protein n=1 Tax=Clavis... 65 3e-09
UniRef100_B6H3W8 Pc13g11020 protein n=1 Tax=Penicillium chrysoge... 65 3e-09
UniRef100_A7EJV5 Putative uncharacterized protein n=1 Tax=Sclero... 65 3e-09
UniRef100_Q0I7K2 Translation initiation factor IF-2 n=1 Tax=Syne... 65 3e-09
UniRef100_Q6ZCX3 Formin-like protein 6 n=1 Tax=Oryza sativa Japo... 65 3e-09
UniRef100_UPI0001B44C51 PE-PGRS family protein n=1 Tax=Mycobacte... 64 4e-09
UniRef100_UPI0001A5EE3F PREDICTED: hypothetical protein FLJ22184... 64 4e-09
UniRef100_UPI0001A5EAAB PREDICTED: hypothetical protein LOC80164... 64 4e-09
UniRef100_UPI0001951234 Ras-associated and pleckstrin homology d... 64 4e-09
UniRef100_B2HFK5 PE-PGRS family protein, PE_PGRS9 n=1 Tax=Mycoba... 64 4e-09
UniRef100_B0TGJ1 Putative uncharacterized protein n=1 Tax=Heliob... 64 4e-09
UniRef100_A5U5I6 PE-PGRS family protein n=3 Tax=Mycobacterium tu... 64 4e-09
UniRef100_C6DMZ3 Putative uncharacterized protein n=1 Tax=Mycoba... 64 4e-09
UniRef100_A5WQA0 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 64 4e-09
UniRef100_Q8GTL0 Putative glycine-rich cell wall protein n=2 Tax... 64 4e-09
UniRef100_Q49I34 120 kDa pistil extensin-like protein (Fragment)... 64 4e-09
UniRef100_Q39789 Proline-rich cell wall protein n=1 Tax=Gossypiu... 64 4e-09
UniRef100_Q39763 Proline-rich cell wall protein n=1 Tax=Gossypiu... 64 4e-09
UniRef100_B6SVB0 Receptor protein kinase PERK1 n=1 Tax=Zea mays ... 64 4e-09
UniRef100_B3XYC5 Chitin-binding lectin n=1 Tax=Solanum lycopersi... 64 4e-09
UniRef100_A9TLH5 Predicted protein (Fragment) n=1 Tax=Physcomitr... 64 4e-09
UniRef100_A8HMN2 Predicted protein n=1 Tax=Chlamydomonas reinhar... 64 4e-09
UniRef100_Q7SFQ1 Predicted protein n=1 Tax=Neurospora crassa Rep... 64 4e-09
UniRef100_Q2KFY0 Putative uncharacterized protein n=1 Tax=Magnap... 64 4e-09
UniRef100_A8NN84 Putative uncharacterized protein n=1 Tax=Coprin... 64 4e-09
UniRef100_A4RBS7 Putative uncharacterized protein n=1 Tax=Magnap... 64 4e-09
UniRef100_UPI0001A7B0C1 protein binding / structural constituent... 64 5e-09
UniRef100_UPI0000E203DD PREDICTED: mucin 7, salivary isoform 2 n... 64 5e-09
UniRef100_UPI00016E8885 UPI00016E8885 related cluster n=1 Tax=Ta... 64 5e-09
UniRef100_Q7VEL2 Putative uncharacterized protein PE_PGRS37 n=1 ... 64 5e-09
UniRef100_Q0S0X8 Putative uncharacterized protein n=1 Tax=Rhodoc... 64 5e-09
UniRef100_B2HSI7 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 64 5e-09
UniRef100_A5U2F3 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 64 5e-09
UniRef100_A0PQV1 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 64 5e-09
UniRef100_C6DPT5 PE-PGRS family protein n=5 Tax=Mycobacterium tu... 64 5e-09
UniRef100_C2GDZ6 Possible Fe-S dehydrogenase n=1 Tax=Corynebacte... 64 5e-09
UniRef100_A5WPA0 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 64 5e-09
UniRef100_Q41805 Extensin-like protein n=1 Tax=Zea mays RepID=Q4... 64 5e-09
UniRef100_Q39353 Cell wall-plasma membrane linker protein n=1 Ta... 64 5e-09
UniRef100_B6SUZ1 Putative uncharacterized protein n=1 Tax=Zea ma... 64 5e-09
UniRef100_Q657S4 Os01g0110200 protein n=2 Tax=Oryza sativa RepID... 64 5e-09
UniRef100_Q5CLM1 Putative uncharacterized protein n=1 Tax=Crypto... 64 5e-09
UniRef100_Q57ZV5 Mitochondrial DNA polymerase beta-PAK n=1 Tax=T... 64 5e-09
UniRef100_B4NC76 GK25807 n=1 Tax=Drosophila willistoni RepID=B4N... 64 5e-09
UniRef100_B4MN10 GK16584 n=1 Tax=Drosophila willistoni RepID=B4M... 64 5e-09
UniRef100_B4IVL8 GE14975 n=1 Tax=Drosophila yakuba RepID=B4IVL8_... 64 5e-09
UniRef100_B3M929 GF25073 n=1 Tax=Drosophila ananassae RepID=B3M9... 64 5e-09
UniRef100_A9URP0 Predicted protein n=1 Tax=Monosiga brevicollis ... 64 5e-09
UniRef100_A7RPE6 Predicted protein n=1 Tax=Nematostella vectensi... 64 5e-09
UniRef100_Q6CCD3 YALI0C10450p n=1 Tax=Yarrowia lipolytica RepID=... 64 5e-09
UniRef100_Q55S44 Putative uncharacterized protein n=1 Tax=Filoba... 64 5e-09
UniRef100_B6QR64 Proline-rich, actin-associated protein Vrp1, pu... 64 5e-09
UniRef100_P14918 Extensin n=1 Tax=Zea mays RepID=EXTN_MAIZE 64 5e-09
UniRef100_UPI0001B4C6CF beta-ketoacyl synthase n=1 Tax=Streptomy... 64 7e-09
UniRef100_UPI0001B46966 PE-PGRS family protein n=1 Tax=Mycobacte... 64 7e-09
UniRef100_UPI00017C2F7A PREDICTED: hypothetical protein n=1 Tax=... 64 7e-09
UniRef100_UPI00015B42DB PREDICTED: hypothetical protein n=1 Tax=... 64 7e-09
UniRef100_UPI0000DA2535 PREDICTED: hypothetical protein n=1 Tax=... 64 7e-09
UniRef100_UPI0000E7F7FF PREDICTED: similar to N-WASP n=1 Tax=Gal... 64 7e-09
UniRef100_A5U298 PE-PGRS family protein n=2 Tax=Mycobacterium tu... 64 7e-09
UniRef100_Q8GFF2 Putative uncharacterized protein n=1 Tax=Strept... 64 7e-09
UniRef100_C6DTI7 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 64 7e-09
UniRef100_C5EUN4 Predicted protein n=1 Tax=Clostridiales bacteri... 64 7e-09
UniRef100_C2A7G2 Putative uncharacterized protein n=1 Tax=Thermo... 64 7e-09
UniRef100_A5WM79 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 64 7e-09
UniRef100_Q9XIV1 Arabinogalactan protein n=1 Tax=Cucumis sativus... 64 7e-09
UniRef100_Q5VMY9 Hydroxyproline-rich glycoprotein-like n=1 Tax=O... 64 7e-09
UniRef100_O22514 Proline rich protein n=1 Tax=Santalum album Rep... 64 7e-09
UniRef100_C4JA68 Putative uncharacterized protein n=1 Tax=Zea ma... 64 7e-09
UniRef100_C4IZA5 Putative uncharacterized protein n=1 Tax=Zea ma... 64 7e-09
UniRef100_B9T5W9 ATP binding protein, putative n=1 Tax=Ricinus c... 64 7e-09
UniRef100_B9RZI2 Protein binding protein, putative n=1 Tax=Ricin... 64 7e-09
UniRef100_A4S5W2 Predicted protein n=1 Tax=Ostreococcus lucimari... 64 7e-09
UniRef100_A2YQ51 Putative uncharacterized protein n=1 Tax=Oryza ... 64 7e-09
UniRef100_A1KR25 Cell wall glycoprotein GP2 (Fragment) n=2 Tax=C... 64 7e-09
UniRef100_C9ZJL6 Putative uncharacterized protein (Fragment) n=1... 64 7e-09
UniRef100_B9PLD5 Putative uncharacterized protein n=1 Tax=Toxopl... 64 7e-09
UniRef100_Q5KG31 Putative uncharacterized protein n=1 Tax=Filoba... 64 7e-09
UniRef100_C4JPD7 Putative uncharacterized protein n=1 Tax=Uncino... 64 7e-09
UniRef100_Q95JC9-2 Isoform 2 of Basic proline-rich protein n=1 T... 64 7e-09
UniRef100_UPI0001924514 PREDICTED: hypothetical protein, partial... 63 9e-09
UniRef100_UPI0000E82544 PREDICTED: similar to alpha-NAC, muscle-... 63 9e-09
UniRef100_UPI000023CA6C hypothetical protein FG00996.1 n=1 Tax=G... 63 9e-09
UniRef100_UPI000162C860 nascent polypeptide-associated complex a... 63 9e-09
UniRef100_UPI00016E9EF6 UPI00016E9EF6 related cluster n=1 Tax=Ta... 63 9e-09
UniRef100_UPI00016E98C3 UPI00016E98C3 related cluster n=1 Tax=Ta... 63 9e-09
UniRef100_Q9Q5L3 EBNA-2 n=1 Tax=Macacine herpesvirus 4 RepID=Q9Q... 63 9e-09
UniRef100_A1XYV9 Latency-associated nuclear antigen n=1 Tax=Retr... 63 9e-09
UniRef100_A8IEX2 Putative uncharacterized protein n=1 Tax=Azorhi... 63 9e-09
UniRef100_A4F5Y5 Putative uncharacterized protein n=1 Tax=Saccha... 63 9e-09
UniRef100_Q9SX31 F24J5.8 protein n=1 Tax=Arabidopsis thaliana Re... 63 9e-09
UniRef100_Q4U2V7 Hydroxyproline-rich glycoprotein GAS31 n=1 Tax=... 63 9e-09
UniRef100_Q39367 Glycine-rich protein (Fragment) n=1 Tax=Brassic... 63 9e-09
UniRef100_Q09084 Extensin (Class II) n=1 Tax=Solanum lycopersicu... 63 9e-09
UniRef100_O49986 120 kDa style glycoprotein n=1 Tax=Nicotiana al... 63 9e-09
UniRef100_B9GF15 Predicted protein n=1 Tax=Populus trichocarpa R... 63 9e-09
UniRef100_B9G7A3 Putative uncharacterized protein n=1 Tax=Oryza ... 63 9e-09
UniRef100_B6T034 Transcriptional regulatory protein algP n=1 Tax... 63 9e-09
UniRef100_A5AN80 Putative uncharacterized protein n=1 Tax=Vitis ... 63 9e-09
UniRef100_Q9VZC2 CG15021 n=1 Tax=Drosophila melanogaster RepID=Q... 63 9e-09
UniRef100_C9ZJL7 Putative uncharacterized protein (Fragment) n=1... 63 9e-09
UniRef100_B7PKI2 Putative uncharacterized protein (Fragment) n=1... 63 9e-09
UniRef100_A9URA4 Predicted protein n=1 Tax=Monosiga brevicollis ... 63 9e-09
UniRef100_A0CZ14 Chromosome undetermined scaffold_31, whole geno... 63 9e-09
UniRef100_A5YT41 Probable cell surface glycoprotein n=1 Tax=uncu... 63 9e-09
UniRef100_Q7G6K7 Formin-like protein 3 n=1 Tax=Oryza sativa Japo... 63 9e-09
UniRef100_UPI0001A7B186 actin binding n=1 Tax=Arabidopsis thalia... 63 1e-08
UniRef100_UPI000190191F hypothetical protein MtubE_03075 n=1 Tax... 63 1e-08
UniRef100_UPI0001553800 PREDICTED: hypothetical protein n=1 Tax=... 63 1e-08
UniRef100_UPI0000DF06C6 Os02g0456000 n=1 Tax=Oryza sativa Japoni... 63 1e-08
UniRef100_UPI0000DD95B9 Os08g0528700 n=1 Tax=Oryza sativa Japoni... 63 1e-08
UniRef100_UPI0000DA267E Wiskott-Aldrich syndrome-like n=1 Tax=Ra... 63 1e-08
UniRef100_UPI00016E2CCA UPI00016E2CCA related cluster n=1 Tax=Ta... 63 1e-08
UniRef100_Q7TPN5 Wiskott-Aldrich syndrome-like (Human) n=2 Tax=M... 63 1e-08
UniRef100_Q9RVT4 Putative uncharacterized protein n=1 Tax=Deinoc... 63 1e-08
UniRef100_B8JAJ3 General secretory system II protein E domain pr... 63 1e-08
UniRef100_B2HRI5 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 63 1e-08
UniRef100_B2HP28 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 63 1e-08
UniRef100_B2HJP2 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 63 1e-08
UniRef100_B2HI03 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 63 1e-08
UniRef100_A3Q2Q5 Putative uncharacterized protein n=1 Tax=Mycoba... 63 1e-08
UniRef100_C6DV78 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 63 1e-08
UniRef100_C6DQY9 PE-PGRS family protein n=1 Tax=Mycobacterium tu... 63 1e-08
UniRef100_Q9LV48 Protein kinase-like protein n=1 Tax=Arabidopsis... 63 1e-08
UniRef100_Q94JZ6 At3g24600 n=1 Tax=Arabidopsis thaliana RepID=Q9... 63 1e-08
UniRef100_Q6QJ28 Putative uncharacterized protein (Fragment) n=1... 63 1e-08
UniRef100_Q43682 Extensin-like protein (Fragment) n=1 Tax=Vigna ... 63 1e-08
UniRef100_C5X5H9 Putative uncharacterized protein Sb02g042660 n=... 63 1e-08
UniRef100_C5WP66 Putative uncharacterized protein Sb01g026500 n=... 63 1e-08
UniRef100_B9F5L5 Putative uncharacterized protein n=1 Tax=Oryza ... 63 1e-08
UniRef100_A9P9Z1 Predicted protein n=1 Tax=Populus trichocarpa R... 63 1e-08
UniRef100_Q5CRN0 Large low complexity protein with proline/alani... 63 1e-08
UniRef100_A0D1C0 Chromosome undetermined scaffold_34, whole geno... 63 1e-08
UniRef100_B6Q311 Actin associated protein Wsp1, putative n=1 Tax... 63 1e-08
UniRef100_B2W6F1 Putative uncharacterized protein n=1 Tax=Pyreno... 63 1e-08
UniRef100_O08816 Neural Wiskott-Aldrich syndrome protein n=1 Tax... 63 1e-08
UniRef100_Q91YD9 Neural Wiskott-Aldrich syndrome protein n=3 Tax... 63 1e-08
UniRef100_P21997 Sulfated surface glycoprotein 185 n=1 Tax=Volvo... 63 1e-08
UniRef100_Q9LVN1 Formin-like protein 13 n=1 Tax=Arabidopsis thal... 63 1e-08
UniRef100_UPI000198522B PREDICTED: hypothetical protein n=1 Tax=... 62 2e-08
UniRef100_UPI000186B3FA hypothetical protein BRAFLDRAFT_112557 n... 62 2e-08
UniRef100_UPI0000F2E473 PREDICTED: similar to WASL protein n=1 T... 62 2e-08
UniRef100_UPI0000E49E39 PREDICTED: similar to Wiskott-Aldrich sy... 62 2e-08
UniRef100_UPI0000E49669 PREDICTED: similar to Wiskott-Aldrich sy... 62 2e-08
UniRef100_UPI0000DF08BC Os02g0794900 n=1 Tax=Oryza sativa Japoni... 62 2e-08
UniRef100_UPI00015A6C38 UPI00015A6C38 related cluster n=1 Tax=Da... 62 2e-08
UniRef100_UPI000069F08E UPI000069F08E related cluster n=1 Tax=Xe... 62 2e-08
UniRef100_UPI000069DD7A UPI000069DD7A related cluster n=1 Tax=Xe... 62 2e-08
UniRef100_UPI0000D667F5 sterile alpha motif domain containing 1 ... 62 2e-08
UniRef100_Q7ZYY2 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 62 2e-08
UniRef100_Q6NYX7 Wiskott-Aldrich syndrome, like n=1 Tax=Danio re... 62 2e-08
UniRef100_Q7U0P7 PE-PGRS FAMILY PROTEIN n=1 Tax=Mycobacterium bo... 62 2e-08
UniRef100_Q3SNT6 OmpA/MotB n=1 Tax=Nitrobacter winogradskyi Nb-2... 62 2e-08
UniRef100_Q21D92 OmpA/MotB n=1 Tax=Rhodopseudomonas palustris Bi... 62 2e-08
UniRef100_B2HT93 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 62 2e-08
UniRef100_B2HQU6 PE-PGRS family protein n=1 Tax=Mycobacterium ma... 62 2e-08
UniRef100_A9GSV1 Putative uncharacterized protein n=1 Tax=Sorang... 62 2e-08
UniRef100_A1KKK5 PE-PGRS family protein n=3 Tax=Mycobacterium bo... 62 2e-08
UniRef100_A0PUN3 PE-PGRS family protein n=1 Tax=Mycobacterium ul... 62 2e-08
UniRef100_Q8VK17 PE_PGRS family protein n=1 Tax=Mycobacterium tu... 62 2e-08
UniRef100_Q9SPM1 Extensin-like protein n=1 Tax=Solanum lycopersi... 62 2e-08
UniRef100_Q5JNC9 Os01g0738300 protein n=1 Tax=Oryza sativa Japon... 62 2e-08
UniRef100_Q2QVX3 Transposon protein, putative, CACTA, En/Spm sub... 62 2e-08
UniRef100_Q00ZZ2 Membrane coat complex Retromer, subunit VPS5/SN... 62 2e-08
UniRef100_C5YH07 Putative uncharacterized protein Sb07g003820 n=... 62 2e-08
UniRef100_C5XIZ1 Putative uncharacterized protein Sb03g013310 n=... 62 2e-08
UniRef100_C5WNL2 Putative uncharacterized protein Sb01g038690 n=... 62 2e-08
UniRef100_C0P2F3 Putative uncharacterized protein n=1 Tax=Zea ma... 62 2e-08
UniRef100_B9RL99 Putative uncharacterized protein n=1 Tax=Ricinu... 62 2e-08
UniRef100_B9EZL1 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_B8A933 Putative uncharacterized protein n=1 Tax=Oryza ... 62 2e-08
UniRef100_B4LE96 GJ13031 n=1 Tax=Drosophila virilis RepID=B4LE96... 62 2e-08
UniRef100_B4IVG9 GE14936 n=1 Tax=Drosophila yakuba RepID=B4IVG9_... 62 2e-08
UniRef100_B4IPI2 GM11753 (Fragment) n=1 Tax=Drosophila sechellia... 62 2e-08
UniRef100_C9J285 Putative uncharacterized protein ENSP0000039240... 62 2e-08
UniRef100_Q6AHT0 Protease-1 (PRT1) protein, putative n=1 Tax=Pne... 62 2e-08
UniRef100_Q15637-5 Isoform 5 of Splicing factor 1 n=1 Tax=Homo s... 62 2e-08
UniRef100_P27483 Glycine-rich cell wall structural protein n=1 T... 62 2e-08